Abstract
Background
In the study of biomolecular structures and interactions the polar hydrogen-π bonds (Hp-π) are an extensive molecular interaction type. In proteins 11 of 20 natural amino acids and in DNA (or RNA) all four nucleic acids are involved in this type interaction.
Results
The Hp-π in proteins are studied using high level QM method CCSD/6-311 + G(d,p) + H-Bq (ghost hydrogen basis functions) in vacuum and in solutions (water, acetonitrile, and cyclohexane). Three quantum chemical methods (B3LYP, CCSD, and CCSD(T)) and three basis sets (6-311 + G(d,p), TZVP, and cc-pVTZ) are compared. The Hp-π donors include R2NH, RNH2, ROH, and C6H5OH; and the acceptors are aromatic amino acids, peptide bond unit, and small conjugate π-groups. The Hp-π interaction energies of four amino acid pairs (Ser-Phe, Lys-Phe, His-Phe, and Tyr-Phe) are quantitatively calculated.
Conclusions
Five conclusion points are abstracted from the calculation results. (1) The common DFT method B3LYP fails in describing the Hp-π interactions. On the other hand, CCSD/6-311 + G(d,p) plus ghost atom H-Bq can yield better results, very close to the state-of-the-art method CCSD(T)/cc-pVTZ. (2) The Hp-π interactions are point to π-plane interactions, possessing much more interaction conformations and broader energy range than other interaction types, such as common hydrogen bond and electrostatic interactions. (3) In proteins the Hp-π interaction energies are in the range 10 to 30 kJ/mol, comparable or even larger than common hydrogen bond interactions. (4) The bond length of Hp-π interactions are in the region from 2.30 to 3.00 Å at the perpendicular direction to the π-plane, much longer than the common hydrogen bonds (~1.9 Å). (5) Like common hydrogen bond interactions, the Hp-π interactions are less affected by solvation effects.

Similar content being viewed by others
Background
The structures of proteins and other biological molecules are determined by the delicate balance between several molecular interactions [1–3]. Among them hydrogen-π interactions [4–7] are an extensive interaction type in organic and biological molecules, referring to the interactions between hydrogen atoms, attaching to different atomic groups, and aromatic molecules or π-groups. The hydrogen-π interactions can be classified into two groups, the nonpolar hydrogen-π interactions (H-π or CH-π) [8–11] and the polar hydrogen-π interactions (Hp-π) [12–16]. The typical nonpolar hydrogen-π interactions are the interactions between hydrogen atoms, attaching to carbon atoms, and the conjugate π-systems, often indicated by the notation CH-π in references [8, 11]. The interaction strength and the physical nature and properties of nonpolar H-π interactions are often a debatable topic [6, 10, 17]. In the polar hydrogen-π interactions the donors are the polar hydrogen atoms, attaching to electronegative atoms (R2NH, RNH2, and ROH), and the acceptors are various aromatic molecules and conjugate π-groups [18–20]. The interaction energies of polar hydrogen-π interactions are much stronger than that of nonpolar hydrogen-π interactions (CH-π), comparable or even larger than common hydrogen bonds. In this study the notation Hp-π is used for the polar hydrogen-π interactions, to make difference from the nonpolar hydrogen-π interactions (H-π or CH-π), and the common hydrogen bond interactions (H-b) [21–25].
The Hp-π interactions frequently happen in biological macromolecules, such as proteins and DNA (or RNA), and play important roles in protein structures and biological functions. In proteins the Hp-π interaction donors are the polar hydrogen atoms in atomic groups [12–16] –NH2, >NH, –OH, –SH, and C6H5OH, which exist in amino acids Ser, Thr, Tyr, Trp, Cys, His, Asn, Gln, Lys, and Arg, while the acceptors are the aromatic amino acids, Phe, Tyr, Trp, and His [19, 20]. Three amino acids play the roles of both donor and acceptor (His, Tyr, and Trp). In the 20 natural amino acids 11 of them may be involved in the Hp-π interactions. The peptide bond units are a quasi π-group, comprising N, C, and O, which can play the roles of both Hp-π interaction acceptor and donor [6]. In DAN and RNA the four nucleic acid components (adenine, guanine, cytosine, and thymine) possess aromatic rings and polar hydrogen groups, which are the Hp-π interaction acceptors and donors [23].
The Hp-π interactions are a unique interaction type that cannot be classified into other molecular interaction types, such as common hydrogen bond [21–25], cation-π interaction [26–29], electrostatic interaction, and van der Waals interaction. The physical nature and the interaction strength of Hp-π interactions are often a debatable topic. In the Hp-π interactions electron dispersion energy is one of the main contributions. Different quantum chemical methods may give very different interaction energies and descriptions for the interaction properties of Hp-π interactions. One reason is that some quantum chemical methods and basis sets may fail in estimating the dispersion energies accurately [30]. In this study several quantum chemical methods and basis sets are evaluated and compared in the Hp-π interaction calculations. The Hp-π interaction contributors, investigated in this study, include the interaction donors R2NH, RNH2, ROH, C6H5OH, and peptide bond unit NMA (n-methyl acetamide); and the interaction acceptors include aromatic molecular benzene, hetero-aromatic molecules C5H5N and imidazole, peptide bond unit, carbonyl group, and other small conjugate π-groups.
Results
In this section all calculation results are summarized and reported using tables and figures. Brief comparisons and illustrations are provided following the results.
Hp-π interactions of small conjugate π-groups
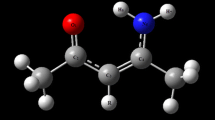
In this section the Hp-π interactions between small donors and acceptors are used to illustrate the Hp-π interaction properties and strength, including three Hp-π interaction pairs CH3OH–H2CO, NMA–C2H4, and CH3OH–C2H4. The Hp-π interaction structures of three Hp-π interaction pairs are shown in Figure 1, and the Hp-π interaction energies are listed in Table 1. In Figure 1 the small blue circle are ‘ghost’ hydrogen atoms, which improve the Hp-π QM calculations effectively. The role of ‘ghost’ atoms will be introduced in detail in the Method section. The HOMOs (highest occupied molecular orbitals) of three Hp-π interaction pairs are shown in Figure 1, in which the polar hydrogen atoms are in close touching with the π-orbitals of the aromatic molecules. In other words, in the Hp-π interactions the polar hydrogen atoms are buried in the π-electron density of aromatic molecules, similar to the common hydrogen bond interactions.
The Hp-π interaction structures and HOMOs of three small interaction pairs. (A) Structure of Hp-π interaction pair CH3OH-H2CO. The polar hydrogen atom of CH3OH points to the carbon atom of H2CO perpendicularly. (B) The HOMO of Hp-π interaction pair CH3OH-H2CO. The polar hydrogen atom is in close touching with the π-orbital of H2CO. (C) Structure of Hp-π interaction pair NMA-C2H4. The polar hydrogen atom of NMA (n-methyl acetamide) points to the center of double bond. (D) The HOMO of Hp-π interaction pair NMA-C2H4. (E) Structure of Hp-π interaction pair CH3OH-C2H4. The polar hydrogen atom of CH3OH points to the center of double bond perpendicularly. (F) The HOMO of Hp-π interaction pair CH3OH-C2H4. The ghost hydrogen atom H-Bq is attached to the polar hydrogen atoms, and the distance to polar hydrogen is 0.90 Å.
Unlike common hydrogen bond interactions that are point-to-point interactions, the Hp-π interactions are point to π-group interactions, in which the interactions could happen at any position of the π-plane. In Table 1 for each Hp-π interaction pair two or more interaction positions are reported. In the NMA–C2H4 interaction pair the Hp-π energy at the bond center (-12.885 kJ/mol) is higher than that (-11.296 kJ/mol) at the carbon atom of C2H4. The structural conformations of Hp-π interactions are much more than that of the common hydrogen bond, and the Hp-π interaction energies could change in a broad range. The Hp-π interaction energies of all three small interaction pairs are larger than half of H2O–H2O hydrogen bond. The NMA (n-methyl acetamide) [31–33] is used as the model of peptide bond units. The Hp-π energy of NMA may represent the Hp-π interactions between the protein peptide backbones and aromatic side chains. The lower part of Table 1 is the Hp-π interactions in three solutions (water, acetonitrile, and cyclohexane) using CCSD and PCM method [34–37]. The Hp-π energies in solutions are decrease with the increase of the solvent dielectric constants ϵ. However, the Hp-π energies in aqueous solution are still large. At this point the Hp-π interactions are like common hydrogen bond interaction, lees affected by solvation effects.
The Hp-π interaction energies as the functions of interaction distances (R) are shown in Figure 2. For comparison the curve of NMA-NMA hydrogen bond interaction is also shown in Figure 2, which is the most frequent hydrogen bond in proteins. The bong lengths (~2.5 Å) of Hp-π interactions are longer than that of the hydrogen bonds (~2.0 Å). The force constants (k) of two Hp-π interaction pairs CH3OH–C2H4 and CH3OH–C6H6 are 0.0035 and 0.0055 Hartree/Bohr, smaller that (0.0071 Hartree/Bohr) of the NMA-NMA hydrogen bond interaction. The Hp-π interactions are more soften than the common hydrogen bond interactions in the minimum and short interaction distances.
The Hp-π interaction energies of CH 3 OH-C 2 H 4 (blue diamond) and CH 3 OH-C 6 H 6 (green square) pairs as the function of interaction distance (R). For comparison the curve of NMA-NMA hydrogen bond interaction is also shown (orange triangles), which is the most frequent hydrogen bond in proteins. The bong lengths (~2.5 Å) of Hp-π interactions are longer than that of the hydrogen bonds (~2.0 Å). The force constants (k) of two Hp-π interaction pairs CH3OH-C2H4 and CH3OH-C6H6 are 0.0035 and 0.0055 Hartree/Bohr, smaller that (0.0071 Hartree/Bohr) of the NMA-NMA hydrogen bond interaction. The Hp-π interactions are more soften than the common hydrogen bond interactions in the minimum and short interaction distances.
Hp-π interactions in aromatic molecules
The aromatic molecules are the best H-π interaction acceptors. In this section the Hp-π interactions of two aromatic molecules are studied. One is the typical aromatic molecule benzene, and the other is a heteroatom-aromatic molecule C5H5N. The interaction structures of three interaction pairs (CH3OH–C6H6, CH3OH–C5H5N, and NMA–C6H6) are shown in Figure 3, and the interaction energies and bond lengths are summarized in Table 2.
The Hp-π interaction structures and HOMOs of three aromatic interaction pairs. (A) Structure of Hp-π interaction pair CH3OH-C6H6. The polar hydrogen atom of CH3OH points to the center of C6H6 perpendicularly. (B) The HOMO of Hp-π interaction pair CH3OH-C6H6. The polar hydrogen atom is in close touching with the π-orbital of C6H6. (C) Structure of Hp-π interaction pair CH3OH-C5H5N. The polar hydrogen atom of CH3OH points to the N of C5H5N. (D) The HOMO of Hp-π interaction pair CH3OH-C5H5N. (E) Structure of Hp-π interaction pair NMA-C6H6. The polar hydrogen atom of NMA (n-methyl acetamide) points to the center of C6H6 perpendicularly. (F) The HOMO of Hp-π interaction pair NMA-C6H6.
The H-π interaction energies of benzene are remarkably larger than that of the small Hp-π interaction acceptors (C2H2 and H2CO) listed in Table 1. Usually the H-π interaction energies increase with the size of aromatic molecules. The Hp-π interaction energy (-14.853 kJ/mol) of heteroatom aromatic molecule (C5H5N) is smaller than the pure aromatic molecule C6H6 (-22.233 kJ/mol). The Hp-π interaction energy of NMA–C6H6 (-22.233 kJ/mol) is comparable to the H2O-H2O hydrogen bond energy (-17 ~ -23 kJ/mol) [38]. This value could represent the Hp-π interaction energies between peptide bond units and the aromatic amino acids in proteins.
Hp-π interactions in amino acids
In the 20 natural amino acids 4 of them possess aromatic side chains (Phe, Tyr, Trp, and His), which are the possible Hp-π interaction acceptors. On the other hand, 10 (Ser, Thr, Asn, Gln, Cys, Tyr, Trp, His, Lys, and Arg) amino acids possess various Hp-π donors. The atomic group –OH is the Hp-π donor in amino acids Ser, Thr, and Tyr. The atomic group > NH or –NH2 is the Hp-π donor of amino acids Asn, Gln, Lys, Arg, Trp, and His. Amino acid Cys has the Hp-π donor –SH. Three amino acids (Tyr, Trp, and His) play the roles of both Hp-π donor and acceptor. In proteins total 11 amino acids are involved in the Hp-π interactions. The Hp-π interactions of amino acids in proteins are an interesting and important research topic. The amino acid Hp-π donors and acceptors are shown in Figure 4, in which the Hp-π donors are indicated using blue cycles, and the Hp-π acceptors are indicated using red cycles.
The Hp-π donors and acceptors of natural amino acids. In the 20 natural amino acids 4 of them possess aromatic side chains (Phe, Tyr, Trp, and His), which are the possible Hp-π interaction acceptors. On the other hand, 10 amino acids (Ser, Thr, Asn, Gln, Cys, Tyr, Trp, His, Lys, and Arg) possess various Hp-π donors. The atomic group –OH is the Hp-π donor in Ser, Thr, and Tyr. The atomic group > NH or –NH2 is the Hp-π donor of Asn, Gln, Lys, Arg, Trp, and His. Amino acid Cys has the donor –SH. Three amino acids (Tyr, Trp, and His) play the roles of both Hp-π donor and acceptor. In proteins total 11 amino acids may be involved in the Hp-π interactions. The Hp-π donors are indicated using blue cycles, and the Hp-π acceptors are indicated using red cycles.
In this section the Hp-π interactions of 4 amino acid pairs (Ser–Phe, Lys–Phe, Tyr–Phe, and His–Phe) are studied and reported. The energies and bond lengths of the four amino acid Hp-π interaction pairs are listed in Table 3, and the interaction geometries are shown in Figure 5. In the calculations the aromatic amino acid Phe is simplified as benzene C6H6, which is the Hp-π acceptor in the four interaction pairs. The four Hp-π donors Ser, Lys, Tyr, and His are simplified as CH3OH, CH3NH2, C6H5OH, and imidazole, respectively, as shown in Figure 5. Except the Lys–Phe pair, the Hp-π interaction energies of other three amino acid pairs are close or even larger than the H2O-H2O hydrogen bond energy. In Table 3 the bond length 3.550 Å of Lys–Phe is from N of Lys to the benzene ring center of Phe, and 3.093 Å is the distance from a polar hydrogen atom of Lys to a carbon atom of Phe. The smaller Hp-π interaction energy (-8.766 kJ/mol) of CH3NH2–C6H6 pair may indicate that the RNH2 is a poorer Hp-π interaction donor than the R2NH (-18.15 kJ/mol of (CH3)2NH–C6H6).
The Hp-π interaction structures and HOMOs of three amino acid interaction pairs. (A) Structure of Hp-π interaction pair CH3NH2-C6H6. The two polar hydrogen atoms of CH3NH2 point to the benzene ring perpendicularly. (B) The HOMO of Hp-π interaction pair CH3NH2-C6H6. The polar hydrogen atoms are in close touching with the π-orbital of C6H6. (C) Structure of Hp-π interaction pair C6H5OH-C6H6. The polar hydrogen atom of C6H5OH points to the center of C6H6. (D) The HOMO of Hp-π interaction pair C6H5OH-C6H6. (E) Structure of Hp-π interaction pair imidazole-C6H6. The polar hydrogen atom of imidazole points to the center of C6H6 perpendicularly. (F) The HOMO of Hp-π interaction pair imidazole-C6H6.
Discussion
In very recently publications [20] the Hp-π interactions were experimentally and theoretically studied by Kumar and Dasa using resonant two photon ionization (R2PI), IR-UV, and UV-UV double resonance spectroscopic techniques, and quantum chemical calculations. In their experiments N–H…π hydrogen bonds, slanted T-shaped structures, were observed in molecular dimer. The experimental observations could be the evidence of Hp-π bonds in molecular interactions.
The polar hydrogen-π (Hp-π) interactions are very different from the non polar hydrogen-π (H-π or CH-π) interactions in interaction strength, and in physical nature and properties. The main physical contributions in Hp-π interactions are the electrostatic interactions, MO coordinating, and electron dispersion interaction. The Hp-π interactions are distance and orientation dependent, and the best orientation is the perpendicular direction from polar hydrogen atom to the π-plane. The interaction energies of polar hydrogen-π interactions (Hp-π) are much stronger than that of the disputed non polar hydrogen-π interactions (CH-π) [8, 9].
The three dimensional structures of proteins are not rigid constructions, but dynamically flexible. The Hp-π interactions are point to π-plane interactions, possessing much more interaction conformations and broader energy range than that of the common hydrogen bond interactions. The Hp-π interactions may be responsible for the flexibility and dynamic activity of proteins. In biological molecules the polar hydrogen atoms are the common interaction donors of both hydrogen bond interactions and Hp-π interactions. These two interaction types happen at different directions. The most favorable orientation (perpendicular direction) for the Hp-π interactions is just the most unfavorable direction for common hydrogen bond interactions. Consequently we can expect that the common hydrogen bond (H-b) interactions and the polar hydrogen-π (Hp-π) interactions are the two main interaction forces supporting the three dimensional structures of proteins, and playing their roles in different directions.
Based on our calculations, the Hp-π energies in solutions are decrease with the increase of the solvent dielectric constants ϵ. In this study the Hp-π interaction energies in solutions are calculated using PCM (Polarized Continuum Model) method. The PCM is a continuing medium model, which cannot give very accurate results. The Hp-π energies in aqueous solution are still significant. At this point the Hp-π interactions are like common hydrogen bond interaction, lees affected by solvation effects. The Hp-π interaction acceptors (aromatic groups) are often the hydrophilic groups, which are buried in the hydrophobic core of protein structures. In the hydrophobic pockets of proteins the solvent dielectric constants are small, and the Hp-π interactions may be still working well.
Conclusion
The polar hydrogen-π interactions (Hp-π) are a unique interaction type different from other main molecular interaction types in physical nature and properties. Some useful conclusions can be refined as follows. (1) In the 20 natural amino acids 11 of them are involved in the Hp-π interactions, including Ser, Thr, Gln, Asn, Arg, Lys, Phe, Tyr, Trp, Cys, and His. In proteins the Hp-π interaction donors are the atomic groups –NH2, >NH, –OH, –SH, and C6H5OH; while the Hp-π interaction acceptors are various aromatic and heteroatom aromatic groups in amino acid side chains. (2) The peptide bond units in protein backbones are quasi π-bonds, possessing both polar hydrogen atoms and conjugate π-groups, playing the roles of both Hp-π interaction acceptors and donors. (3) The Hp-π interactions are point to π-plane interactions, having many possible interaction conformations. (4) The Hp-π interaction energies between amino acids are in the range from -10 to -25 kJ/mol, close or even larger than the common hydrogen bonds. (5) The bond length of Hp-π interactions are in the region from 2.30 to 3.00 Å at the perpendicular direction to the π-plane, much longer than common hydrogen bonds. (6) Like common hydrogen bond interactions, the Hp-π interactions are less affected by solvation effects. (7) The common DFT method B3LYP fails in describing the Hp-π interactions. On the other hand, CCSD/6-311 + G(d,p) plus ghost atom H-Bq at bond middle can yield better results using less cpu-time, very close to the state-of-the-art method CCSD(T)/cc-pVTZ.
Method and theory
DFT method B3LYP has been widely used in the studies of organic molecules and biological molecules for many years because of its higher accuracy and less calculation workload. However, in recent decade the common DFT methods were found failing in description of molecular dispersion interactions, which are a main contribution in Hp-π interactions. On the other hand, more advanced quantum chemical configuration interaction (CI) [39–41] methods are able to evaluate the dispersion energies. However, such sophisticated methods are expensive, consuming much more CPU times and computer resource than DFT methods do. Particularly the typical bond lengths of Hp-π interactions are around 2.5 Å, much longer than other molecular interaction types, such as hydrogen bonds (~2.0 Å). In order to calculate the long range Hp-π interactions accurately large basis sets have to be used. The large basis sets make the CI calculations of Hp-π interactions are even more expensive and CUP-time consuming. Careful selection of appropriable methods for the Hp-π interaction calculations is the first step of the Hp-π study.
A comprehensive comparison was performed to evaluate the calculation results of several methods, including three methods (B3LYP, CCSD, and CCSD(T)) [42–45] and three basis sets (6-311 + G(d,p), TZVP, and cc-pVTZ) [46, 47]. Small Hp-π interaction donors (CH3OH) and two acceptors (C2H4 and C6H6) are used in the comparison calculations. The Hp-π interaction energy of CH3OH-C2H4, one of the smallest Hp-π interaction pairs, is -11.853 kJ/mol, more than half of the H2O-H2O hydrogen bond energy -21.258 kJ/mol. The Hp-π interaction energy of CH3OH-C6H6 is -19.765 kJ/mol, very close to the common hydrogen bonds. In biological molecular interactions the Hp-π interactions are comparable to the hydrogen bond interactions. A remarkable difference between Hp-π interactions and H-b interactions is that the hydrogen bonds are point to point interactions, and the Hp-π bonds are point to π-plane interactions.
The Hp-π interaction structures and HOMO (highest occupied molecular orbital) of the Hp-π interaction pairs are shows in Figure 1. In the interaction pair CH3OH-C2H4 the polar hydrogen of CH3OH points to the double bond center of C2H4 perpendicularly, and the polar hydrogen atom points to the carbon and the bond center of C2H4, respectively. In the HOMO figures of CH3OH-C2H4 interaction pair the polar hydrogen atoms are in close touching with the π-orbitals of C2H4. At this point Hp-π interactions are similar to the hydrogen bond interactions. In the latter the polar hydrogen atoms are buried in the electron density of electronegative atoms, such as oxygen and nitrogen.
The more advanced CI method CCSD (T) gives better results than that of the CCSD method. However, the cpu-time of CCSD (T) is much longer than that of the CCSD. Generally speaking the larger basis sets give the better results. In the calculations of Hp-π interactions polarization functions, diffuse functions, and floating functions are necessary. However, large basis set remarkably increases the cpu-time of CCSD (T) calculations. In solving this problem a simple method is the use of ‘ghost atoms’. The ghost hydrogen atom H-Bq is an empty atom possessing the basis functions of hydrogen, but having no nucleus charge and electron [48, 49]. In the CCSD calculation using 6-311 + G(d,p) basis set plus a hydrogen ‘ghost atom’ H-Bq yields the result (-11.715 kJ/mol), very close to the result (-11.853 kJ/mol) of the state-of-the-art method CCSD(T)/cc-pVTZ. However, the cpu-time of CCSD calculation reduces to 1/8 of CCSD(T) calculation (5.3 hours to 44.6 hours). Another advantage of the use of ghost atom is reducing the basis set superposition error (BSSE) [48, 49].
In the Hp-π interaction calculations by using DFT method B3LYP only take cpu-time few minutes. However, the Hp-π interaction energies are around 20 ~ 30% smaller than that of other two CI methods (CCSD and CCSD (T)), because the common DFT methods fail in evaluating the dispersion energies, which is an important contribution in Hp-π interactions. In recent years the shortcoming of DFT methods has been improved by the better density functionals or using empirical correction for dispersion.
In this study all calculations are performed using CCSD method and basis set 6-311 + G(d,p) + H-Bq, in which a ‘ghost hydrogen atom’ (H-Bq) is attached to the polar hydrogen atom, and the distance to polar hydrogen atom is 0.9 Å. Keep in mind, in the calculations for Hp-π interaction energies the same ghost atom is also added to the two molecule monomers. The Hp-π interaction energies in solutions are calculated using CCSD and PCM (Polarized Continuum Model) method [34–37]. All calculations are performed at Sugon-5000A computer and Tianhe-1A computer in National Supper Computing Center in Tianjin (China) using Gaussian 09 and Gauss View 5 software packages [50].
References
Mezey PG: Chemical bonding in proteins and other macromolecules. Theor Comput Chem. 1999, 6: 613-636.
Mezey PG: A crystallographic structure refinement approach using ab initio quality additive fuzzy density fragments. Adv Molec Structure Res. 1998, 4: 115-149.
Szekeres ZS, Mezey PG: Fragmentation Selection Strategies in Linear Scaling Methods. “Linear-Scaling Techniques in Computational Chemistry and Physics, Methods and Applications”. Edited by: Leszczynski J, Zalesny R, Papadopoulos M, Mezey PG. 2011, New York: Springer, 147-156.
Mohan N, Vijayalakshmi KP, Koga N, Suresh CH: Comparison of aromatic NH…π, OH…π, and CH…π interactions of alanine using MP2, CCSD, and DFT Methods. J Comput Chem. 2010, 31: 2874-2882.
Sun C-L, Jiang X-N, Wang C-S: An analytic potential energy function for the amide–amide and amide–water intermolecular hydrogen bonds in peptides. J Comput Chem. 2009, 30: 2567-2575. 10.1002/jcc.21266.
Cheng J, Kang C, Zhu W, Luo X, Puah CM, Chen K, Shen J, Jiang H: N-methylformamide - benzene complex as a prototypical peptide N - H · · · π hydrogen-bonded system: density functional theory and MP2 studies. J Org Chem. 2003, 68: 7490-7495. 10.1021/jo026910b.
Braga D, Grepioni F, Tedesco E: X - H–-π (X = O N C) hydrogen bonds in organometallic crystals. Organometallics. 1998, 17: 2669-2672. 10.1021/om971096h.
Mishra BK, Karthikeyan S, Ramanathan V: Tuning the C–H · · · π interaction by different substitutions in benzene–acetylene complexes. J Chem Theory Comput. 2012, 8: 1935-1942. 10.1021/ct300100h.
Biradha K, Zaworotko MJ: A supramolecular analogue of cyclohexane sustained by aromatic C - H · · · π interactions: Complexes of 135-trihydroxybenzene with substituted pyridines. J Am Chem Soc. 1998, 120: 6431-6432. 10.1021/ja974105m.
Morita S, Fujii A, Mikami N, Tsuzuki S: Origin of the attraction in aliphatic C - H/π interactions: Infrared spectroscopic and theoretical characterization of gas-phase clusters of aromatics with aethane. J Phys Chem A. 2006, 110: 10583-10590. 10.1021/jp064297k.
Birchall LS, Roy S, Jayawarna V, Hughes M, Irvine E, Okorogheye GT, Saudi N, De Santis E, Tuttle T, Edwards AA, Ulijn RV: Exploiting CH-π interactions in supramolecular hydrogels of aromatic carbohydrate amphiphiles. Chem Sci. 2011, 2: 1349-1355. 10.1039/c0sc00621a.
Riley KE, Pitonák M, Cerný J, Hobza P: On the structure and geometry of biomolecular binding motifs (hydrogen-bonding, stacking, X-H · · · π): WFT and DFT calculations. J Chem Theory Comput. 2011, 7: 807-807. 10.1021/ct200019g.
Ren F, Cao D, Wang W, Ren J, Hou S, Chen S: A theoretical study on unusual intermolecular T-shaped X–Hπ interactions between the singlet state HB = BH and HF, HCl, HCN or H2C2. J Mol Model. 2009, 15: 515-523. 10.1007/s00894-008-0415-8.
Liao S-M, Du Q-S, Meng J-Z, Pang Z-W, Huang R-B: The multiple roles of histidine in protein interactions. Chem Central J. 2013, 7: 44-10.1186/1752-153X-7-44.
Grabowski SJ, Lipkowski P: Characteristics of X-H · · · π interactions: Ab initio and QTAIM studies. J Phys Chem A. 2011, 115: 4765-4773. 10.1021/jp2005327.
Ottiger P, Pfaffen C, Leist R, Leutwyler S: Strong N-H…π hydrogen bonding in amide-benzene interactions. J Phys Chem B. 2009, 113: 2937-2943. 10.1021/jp8110474.
Singh NJ, Min SK, Kim DY, Kim KS: Comprehensive energy analysis for various types of π-interaction. J Chem Theory Comput. 2009, 5: 515-529. 10.1021/ct800471b.
Fleming PJ, Rose JD: Do all backbone polar groups in proteins form hydrogen bonds?. Protein Sci. 2005, 14: 1911-1917. 10.1110/ps.051454805.
Meyer EA, Castellano RK, Diederich F: Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed. 2003, 42: 1210-1250. 10.1002/anie.200390319.
Kumar S, Dasa A: Effect of acceptor heteroatoms on π-hydrogen bonding interactions: a study of indole…thiophene heterodimer in a supersonic jet. J Chem Phys. 2012, 137: 094309-10.1063/1.4748818.
Velázquez-Ponce M, Salgado-Zamora H, Jiménez-Vázquez HM, Campos-Aldrete ME, Jiménez R, Cervantes H, Hadda TB: Intramolecular H-bonding interaction in angular 3-pi-EWG substituted imidazo [1,2-a] pyridines contributes to conformational preference. Chem Central J. 2013, 7: 20-10.1186/1752-153X-7-20.
Tabatabaee M: Supramolecular assembled of hexameric water clusters into a 1D chain containing (H2O)6 and [(H2O)4O2] stabilized by hydrogen bonding in a copper complex. Chem Central J. 2012, 6: 5-10.1186/1752-153X-6-5.
Panasik N, Fleming PJ, Rose GD: Hydrogen-bonded turns in proteins: the case for a recount. Protein Sci. 2005, 14: 2910-2914. 10.1110/ps.051625305.
Livesay DR, Huynh DH, Dallakyan S, Jacobs DJ: Hydrogen bond networks determine emergent mechanical and thermodynamic properties across a protein family. Chem Central J. 2008, 2: 17-10.1186/1752-153X-2-17.
Ragone R: Hydrogen-bonding classes in proteins and their contribution to the unfolding reaction. Protein Sci. 2001, 10: 2075-2082. 10.1110/ps.09201.
Du Q-S, Long S-Y, Meng J-Z, Huang R-B: Empirical formulation and parameterization of cation-π interactions for protein modeling. J Comput Chem. 2012, 33: 153-162. 10.1002/jcc.21951.
Du Q-S, Liao S-M, Meng J-Z, Huang R-B: Energies and physicochemical properties of cation-π interactions in biological structures. J Mol Graph Model. 2012, 34: 38-45.
Martis RL, Singh SK, Gromiha MM, Santhosh C: Role of cation–π interactions in single chain ‘all-alpha’ proteins. J Theor Biology. 2008, 250: 655-662. 10.1016/j.jtbi.2007.10.024.
Anbarasu A, Sethumadhavan R: Exploring the role of cation–π interactions in glycoproteins lipid-binding proteins and RNA-binding proteins. J Theor Biology. 2007, 247: 346-353. 10.1016/j.jtbi.2007.02.018.
Du Q-S, Liu P-J, Deng J: Empirical correction to molecular interaction energies in density functional theory (DFT) for methane hydrate simulation. J Chem Theory Comput. 2007, 3: 1665-1672. 10.1021/ct700026d.
Du Q-S, Wei D-Q: Solvation and polarization of the N-methyl amine molecule in aqueous solution: a combined study of quantum mechanics and integral equation theory in three-dimensions. J Phys Chem B. 2003, 107: 13463-13470. 10.1021/jp022493v.
Mannfors B, Mirkin NG, Palmo K, Krimm S: A polarizable electrostatic model of the N-methylacetamide dimer. J Comput Chem. 2001, 22: 1933-1943. 10.1002/jcc.1143.
Cieplak P, Kollman P: On the use of electrostatic potential derived charges in molecular mechanics force fields the relative solvation free energy of cis- and trans-N-methyl-acetamide. J Comput Chem. 1991, 12: 1232-1236. 10.1002/jcc.540121010.
Miertus S, Scrocco E, Tomasi J: Electrostatic interaction of a solute with a continuum a direct utilization of ab initio molecular potentials for the prevision of solvent effects. Chem Phys. 1981, 55: 117-129. 10.1016/0301-0104(81)85090-2.
Amovilli C, Barone V, Cammi R, Cances E, Cossi M, Mennucci B, Pomelli CS, Tomasi J: Recent advances in the description of solvent effects with the polarizable continuum model. Adv Quant Chem. 1998, 32: 227-262.
Cossi M, Barone V: Analytical second derivatives of the free energy in solution by polarizable continuum models. J Chem Phys. 1998, 109: 6246-6254. 10.1063/1.477265.
Foresman JB, Keith TA, Wiberg KB, Snoonian J, Frisch MJ: Solvent effects 5 influence of cavity shape truncation of electrostatics and electron correlation on ab initio reaction field calculations. J Phys Chem. 1996, 100: 16098-16104. 10.1021/jp960488j.
Modig K, Pfrommer BG, Halle B: Temperature dependent Hydrogenbond geometry in liquid water. Phys Rev Lett. 2003, 90: 075502-
Wenzel KB: Configuration interaction (CI): Approximate inclusion of fourfold and threefold excitations, an application of knowledge engineering. J Comput Chem. 1982, 3: 191-207. 10.1002/jcc.540030210.
Sherrill CD, Schaefer HF: The configuration interaction method: advances in highly correlated approaches. Löwdin, Per-Olov Advances in Quantum Chemistry. 1999, San Diego: Academic Press, 143-269. 34
Ma J, Li S, Li W: A multireference configuration interaction method based on the separated electron pair wave functions. J Comput Chem. 2006, 27: 39-47. 10.1002/jcc.20319.
Purvis GD, Bartlett RJ: A full coupled-cluster singles and doubles model: the inclusion of disconnected triples. J Chem Phys. 1982, 76: 1910-1919. 10.1063/1.443164.
Lee TJ, Rice JE: An efficient closed-shell singles and doubles coupled-cluster method. Chem Phys Lett. 1988, 23: 406-415.
Scuseria GE, Schaefer HF: Is coupled cluster singles and doubles (CCSD) more computationally intensive than quadratic configuration interaction (QCISD)?. J Chem Phys. 1989, 90: 3700-3703. 10.1063/1.455827.
Scuseria GE, Janssen CL, Schaefer HF: An efficient reformulation of the closed-shell coupled cluster single and double excitation (CCSD) equations. J Chem Phys. 1988, 89: 7382-7388. 10.1063/1.455269.
Mayer I, Valiron P: Second order Møller–Plesset perturbation theory without basis set superposition error. J Chem Phys. 1998, 109: 3360-3373. 10.1063/1.476931.
Asturioll D, Duran M, Salvador P: Intramolecular basis set superposition error effects on the planarity of benzene and other aromatic molecules: a solution to the problem. J Chem Phys. 2008, 128: 144108-10.1063/1.2902974.
Balabin RM: Enthalpy difference between conformations of normal alkanes: Intramolecular basis set superposition error (BSSE) in the case of n-butane and n-hexane. J Chem Phys. 2008, 129: 164101-10.1063/1.2997349.
Van Duijneveldt FB, van Duijneveldt-van de R, Jeanne GCM, van Lenthe JH: State of the art in counterpoise theory. Chem Rev. 1994, 94: 1873-1885. 10.1021/cr00031a007.
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA: Gaussian 09 Revision B01. 2010, Wallingford: Gaussian Inc
Acknowledgement
This work was supported by the National Science Foundation of China (NSFC) under the grant 31160032, by the Key Natural Science Foundation of Guangxi (GXNSFD) under the grant 2010GXNSFD013030, and by the BaGui Scholar’s Program Foundation. We thank the National Supper Computing Center in Tianjin (NSCC) for the valuable help in the calculations. We appreciate the professional comments and the constructive suggestions of the anonymous reviewers and the editor in improving the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
All authors declare that there are no any competing interests.
Authors’ contributions
All authors contributed equality for the development of the manuscript. RBH and QSD designed the research scheme and wrote the article. QYW did most calculations, LQD and DC performed the data collection and analysis. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Du, QS., Wang, QY., Du, LQ. et al. Theoretical study on the polar hydrogen-π (Hp-π) interactions between protein side chains. Chemistry Central Journal 7, 92 (2013). https://doi.org/10.1186/1752-153X-7-92
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1752-153X-7-92