Abstract
Background
The polymorphism Pro12Ala in peroxisome proliferator-activated receptorγ2 gene (PPARγ2) has been reported to be associated with diabetic nephropathy (DN) in some studies, though the results remain inconclusive. To explore this relationship between PPARγ2 Pro12Ala polymorphism and the susceptibility for DN, a cumulative meta-analysis was performed in this study.
Method
PubMed, Medline, Embase and Web of Science databases have been systematically searched to identify relevant studies. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated.
Results
18 studies were included in this meta-analysis, involving 3,361 cases and 5,815 controls. The PPARγ2 Ala12 allele was significantly associated with decreased risk of DN based on dominant model (OR=0.778; 95%CI=0.618–0.981; Pheterogeneity=0.008; P=0.034). In the stratified analysis by ethnicity, significantly decreased risks were found among Caucasians for dominant model (OR=0.674; 95%CI=0.500–0.909; Pheterogeneity=0.079; P=0.010), while there was no significant association was found in Asians.
Conclusions
The results from the present meta-analysis indicated that the Pro12Ala polymorphism in PPARγ2 gene is not a risk factor for DN in type 2 diabetes (T2D). Further large and well-designed studies are needed to confirm this conclusion.
Virtual slides
The virtual slides for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/7491348341027320.
Similar content being viewed by others
Introduction
Diabetic nephropathy is the leading cause of end-stage renal disease (ESRD) worldwide, and it is estimated that 20% of T2D patients reach ESRD during their lifetime [1]. The pathogenesis of DN has many genetic and environmental factors contributing to its development and progression. The risk of developing DN has been linked to different chromosomes, including chromosome 3, to which the peroxisome proliferators-activated receptor (PPAR) gene has been mapped, particularly to the PPAR gamma (PPARG) nuclear receptor, which is mainly expressed in adipose tissue but is also found in pancreatic beta cells, vascular endothelium, and macrophage [2, 3].
PPAR-γ2 is a nuclear receptor that serves important roles in intermediate metabolism. The PPARG gene is located on chromosome 3p [2]. The Pro12Ala polymorphism of the PPARG gene, a Pro-to-Ala exchange that results in the substitution of proline with alanine at codon 12, was associated with reductions in both DNA binding and transcriptional activity in vitro, and Ala12 carriers showed significant improvement in insulin sensitivity [4, 5]. The Pro12Ala polymorphism in the PPARG gene is suggested to be associated with DN. As we all kwon, many epidemiologic studies investigated the potential association of PPARG gene polymorphisms with susceptibility to DN. However, these studies have reported inconclusive results.
One putative genetic determinant of DN is the Pro12Ala polymorphism in the gene encoding PPARg. PPARg, a member of the nuclear hormone receptor superfamily of ligand-activated transcription factors, plays a key role in regulating the expression of numerous genes involved in lipid metabolism, metabolic syndrome, inflammation, and atherosclerosis [6, 7]. The P12A single-nucleotide polymorphism (SNP) located in the adipocyte-specific PPARg2 isoform has been associated with lower nephropathy in T2D [8, 9]. The underlying molecular mechanism of this polymorphism appears to be, in vitro, a moderate reduction in DNA-binding activity and reduced transcriptional activity [4, 10]. In the present study, we examined the relationship between PPAR-γ2 gene polymorphisms and the risk of T2D DN.
Materials and methods
Search strategy
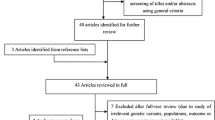
A comprehensive literature search was performed using the PubMed, Medline, Embase, and Web of Science database for relevant articles published (last search updated in May. 2013) with the following key words “PPARγ2”, “polymorphism” and “diabetic nephropathy”. Additional studies were identified by hand searching references in original articles and review articles. Authors were contacted directly regarding crucial data not reported in original articles. The search was limited to human studies. All eligible studies were retrieved, and their bibliographies were checked for other relevant publications. When the same sample was used in several publications, only the most complete study was included following careful examination.
Inclusion criteria
The included studies have to meet the following criteria: (1) only the case–control studies were considered; (2) evaluated the PPARγ2 Pro12Ala polymorphism and DN risk; (3) the genotype distribution of the polymorphism in cases and controls were described in details, and the results were expressed as odds ratio (OR) and corresponding 95% confidence interval (95% CI).
Exclusion criteria
Major reasons for exclusion of studies were as follows: (1) not for DN research; (2) only case population; (3) duplicate of previous publication; and (4) the distribution of genotypes among controls are not in Hardy–Weinberg equilibrium (P <0.01).
Data extraction
Information was carefully extracted from all eligible studies independently by two investigators according to the inclusion criteria listed above. The following data were collected from each study: first author’s name, year of publication, country of origin, ethnicity, source of controls, genotyping method, match, sample size, and numbers of cases and controls in the PPARγ2 Pro12Ala genotypes whenever possible. Ethnicity was categorized as “Caucasian” and “Asian”. When a study did not state which ethnic groups were included or if it was impossible to separate participants according to phenotype, the sample was termed as “mixed population”. We did not define any minimum number of patients to include in this meta-analysis. Articles that reported different ethnic groups and different countries or locations, we considered them different study samples for each category cited above.
Statistical analysis
Crude odds ratios (ORs) together with their corresponding 95% CIs were used to assess the strength of association between the PPARγ2 Pro12Ala polymorphism and DN risk. The pooled ORs were performed for dominant comparison model (Ala-carrier vs Pro/Pro). Between-study heterogeneity was assessed by calculating Q-statistic (Heterogeneity was considered statistically significant if P < 0.10) [11] and quantified using the I2 value, Venice criteria [12] for the I2 test included: “I2< 25% represents no heterogeneity, I2= 25–50% represents moderate heterogeneity, I2= 50–75% represents large heterogeneity, and I2 >75% represents extreme heterogeneity”. If results were not heterogeneous, the pooled ORs were calculated by the fixed-effect model (we used the Q-statistic, which represents the magnitude of heterogeneity between-studies) [13]. Otherwise, a random-effect model was used (when the heterogeneity between-studies were significant) [14]. We also performed subgroup analysis by ethnicity (Caucasian and Asian). Moreover, sensitivity analysis was performed by excluding a single study each time. In addition, we also ranked studies according to sample size, and then repeated this meta-analysis. Begg’s funnel plots [15] and Egger’s linear regression test [16] were used to assess publication bias. All of the calculations were performed using STATA version 12.0 (STATA Corporation, College Station, TX).
Results
Study characteristics
Studies relevant to the searching words were retrieved originally. 18 publications addressing the association between PPARγ2 Pro12Ala polymorphism and DN risk were preliminarily eligible [8, 9, 17–30]. Table 1 presents the main characteristics of these studies. There were a total of 18 studies including 8 groups of Caucasians and 10 groups of Asians. Given the low frequency of Ala allele, even in some studies, Ala homozygous individuals are absent, and only the dominant model was investigated, comparing Ala carriers to Pro/Pro. We also assessed the deviation of Hardy-Weinberg equilibrium in control subjects, and the results demonstrated that all the distribution of genotypes in the controls of all studies was in agreement with Hardy–Weinberg equilibrium.
Meta-analysis results
Overall, the Pro12Ala polymorphism was found to be significantly associated with decreased risk of T2D DN (OR=0.778; 95%CI=0.618–0.981; Pheterogeneity=0.008; P=0.034) (Figure 1). Both the Cochran Q test and estimate of I2 revealed significant heterogeneity among the constituent studies (Pheterogeneity=0.008; I2=50.1%). To avoid the influence of heterogeneity among the included studies, subgroup analyses were distinctively carried out according to the ethnicity. In the stratified analysis by ethnicity, significantly decreased risks were found among Caucasians for dominant model (Ala carrier vs Pro/Pro: OR=0.674; 95%CI=0.500–0.909; Pheterogeneity=0.079; P=0.010), while there was no significant association was found under the same genetic model (OR=0.917; 95%CI=0.639-1.315; Pheterogeneity=0.019; P=0.637) (Figure 2). Significant heterogeneity was found in Asian populations (Pheterogeneity=0.019; I2= 54.7%), but not found in Caucasian populations (Pheterogeneity=0.079; I2=44.9%).
Sensitivity analysis
To test the stability of the pooled results, one-way sensitivity analyses were performed. A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs, no other single study influenced the pooled OR qualitatively, suggesting that the results of this meta-analysis were stable (Figure 3).
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of the literature. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry (PBegg=0.472) (Figure 4). Furthermore, Egger’s test was used to provide statistical evidence for funnel plot symmetry. The results still did not suggest any evidence of publication bias (PEegger=0.234) (Figure 5).
Discussion
It is well recognized that there is individual susceptibility to the DN even with the same environmental exposure. Host factors, including polymorphisms of genes involved in this formation of DN may have accounted for this difference. Therefore, genetic susceptibility to DN has been a research focus in scientific community. Recently, genetic polymorphisms of the PPARγ2 Pro12Ala gene in the etiology of DN have drawn increasing attention. Previous results of the studies on the relationship between PPARγ2 Pro12Ala polymorphisms and DN risk were contradictory. These inconsistent results are possibly because of a small effect of the polymorphism on DN risk or the relatively low statistical power of the published studies. A meta-analysis is a powerful strategy because it potentially investigates a large number of individuals and can estimate the effect of a genetic factor on the risk of the disease [31–35]. To better understanding of this association, a pooled analysis with a large sample, subgroup analysis performed, and heterogeneity explored is necessary to provide a quantitative approach for combining the results of various studies with the same topic, and for estimating the real association in this meta-analysis.
The present study including 3,361 cases and 5,815 controls from 18 published case–control studies, explored the association between a potentially functional polymorphism, PPARγ2 Pro12Ala and T2D DN risk. We found that there was evidence that the variant genotypes of the PPARγ2 were associated with a significant decreased overall risk of DN. When stratified by ethnicity, Caucasians with the Ala carrier showed a decreased risk of DN compared with those with the Pro/Pro genotype. However, Asians did not show the same results.
Some limitations of this meta-analysis should be mentioned. Firstly, bladder cancer is a multifactorial disease that results from complex interactions between many environmental and genetic factors. This means that there will not be single gene or single environmental factor that has large effects on DN susceptibility. Our results were based on unadjusted estimates, while a more precise analysis should be conducted if individual data were available, which would allow for the adjustment by other covariates including age, sex, family history, environmental factors and lifestyle. Secondly, the number of subjects was relatively small, not having enough statistical power to explore the real association. Thirdly, the controls were not uniformly defined. Finally, there were only two ethnic decent populations. So, further large and well-designed studies are needed to confirm this conclusion.
Conclusion
This meta-analysis suggests that the PPARγ2 Pro12Ala polymorphism is not a risk factor for developing T2D DN. Moreover, gene–gene and gene–environment interactions should also be considered in future analysis. Such studies taking these factors into account may eventually lead to our better, comprehensive understanding of the association between the PPARγ2 Pro12Ala polymorphism and T2D DN risk.
Abbreviations
- DN:
-
Diabetic nephropathy
- HWE:
-
Hardy–Weinberg equilibrium
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- PPARγ2:
-
Peroxisome proliferator-activated receptorγ2 gene.
References
Ayodele OE, Alebiosu CO, Salako BL: Diabetic nephropathy: A review of the natural history, burden, risk factors and treatment. J Natl Med Assoc. 2004, 96: 1445-1454.
Moczulski DK, Rogus JJ, Antonellis A, Warram JH, Krolewski AS: Major susceptibility locus for nephropathy in type 1 diabetes on chromosome 3q: Results of novel discordant sib-pair analysis. Diabetes. 1998, 47: 1164-1169. 10.2337/diabetes.47.7.1164.
Guan Y, Zhang Y, Schneider A, Davis L, Breyer RM, Breyer MD: Peroxisome proliferator-activated receptor-gamma activity is associated with renal microvasculature. Am J Physiol Renal Physiol. 2001, 281: F1036-F1046.
Deeb SS, Fajas L, Nemoto M, et al.: A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998, 20: 284-287. 10.1038/3099.
Liu L, Zheng T, Wang F, et al.: Pro12Ala polymorphism in the PPARG gene contributes to the development of diabetic nephropathy in Chinese type 2 diabetic patients. DiabetesCare. 2010, 33: 144-149. 10.2337/dc09-1258.
Argmann CA, Cock TA, Auwerx J: Peroxisome proliferator-activated receptor gamma: The more the merrier?. Eur J Clin Invest. 2005, 35: 82-92. 10.1111/j.1365-2362.2005.01456.x.
Desvergne B, Michalik L, Wahli W: Be fit or be sick: Peroxisome proliferator-activated receptors are down the road. Mol Endocrinol. 2004, 18: 1321-1332. 10.1210/me.2004-0088.
Caramori ML, Canani LH, Costa LA, Gross JL: The human peroxisome proliferator-activated receptor gamma2 (PPAR-gamma2) Pro12Ala polymorphism is associated with decreased risk of diabetic nephropathy in patients with type 2 diabetes. Diabetes. 2003, 52: 3010-3013. 10.2337/diabetes.52.12.3010.
Herrmann SM, Ringel J, Wang JG, Staessen JA, Brand E: Peroxisome proliferator-activated receptor-gamma2 polymorphism Pro12Ala is associated with nephropathy in type 2 diabetes: The Berlin Diabetes Mellitus (BeDiaM) Study. Diabetes. 2002, 51: 2653-2657. 10.2337/diabetes.51.8.2653.
Masugi J, Tamori Y, Mori H, Koike T, Kasuga M: Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-gamma 2 on thiazolidinedione-induced adipogenesis. Biochem Biophys Res Commun. 2000, 268: 178-182. 10.1006/bbrc.2000.2096.
Davey SG, Egger M: Meta-analyses of randomized controlled trials. Lancet. 1997, 350: 1182-
Ioannidis JP, Boffetta P, Little J, et al.: Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008, 37: 120-132. 10.1093/ije/dym159.
Mantel N, Haenszel W: Statistical aspects of the analysis of data from retrospective studies of disease. Natl Cancer Inst. 1959, 22: 719-778.
DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials. 1986, 7: 177-188. 10.1016/0197-2456(86)90046-2.
Begg CB, Mazumdar M: Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994, 50: 1088-1101. 10.2307/2533446.
Egger M, Smith DG, Schneider M, Minder C: Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997, 315: 629-634. 10.1136/bmj.315.7109.629.
Mori H, Ikegami H, Kawaguchi Y, et al.: The Pro12→Ala substitution in PPAR-gamma is associated with resistance to development of diabetes in the general population: possible involvement in impairment of insulin secretion in individuals with type 2 diabetes. Diabetes. 2001, 50: 891-894. 10.2337/diabetes.50.4.891.
Wu S: Study on the Relationship of Polymorphism of PPAR-g2 and Type 2 Diabetes and Diabetic Nephropathy. 2004, Tianjin: Tianjin Medical University
Maeda A, Gohda T, Funabiki K, Horikoshi S, Tomino Y: Peroxisome proliferator-activated receptor gamma gene polymorphism is associated with serum triglyceride levels and body mass index in Japanese type 2 diabetic patients. J Clin Lab Anal. 2004, 18: 317-321. 10.1002/jcla.20045.
Pollex RL, Mamakeesick M, Zinman B, Harris SB, Hegele RA, Hanley AJ: Peroxisome proliferator-activated receptor gamma polymorphism Pro12Ala is associated with nephropathy in type 2 diabetes. J Diabetes Complications. 2007, 21: 166-171. 10.1016/j.jdiacomp.2006.02.006.
Erdogan M, Karadeniz M, Eroglu Z, Tezcanli B, Selvi N, Yilmaz C: The relationship of the peroxisome proliferator-activated receptor-gamma 2 exon 2 and exon 6 gene polymorphism in Turkish type 2 diabetic patients with and without nephropathy. Diabetes Res Clin Pract. 2007, 78: 355-359. 10.1016/j.diabres.2007.06.005.
Wei F, Wang J, Wang C, Huo X, Xue Y, Bai L: The relationship between the pro12Ala polymorphism in PPAR-g2 gene and diabetic nephropathy in Baotou. Chinese J Diabetes. 2008, 16: 679-680.
Li L, Liu L, Zheng T, Wang N, Wang F: Peroxisome proliferator activated receptor g2 gene P12A polymorphism and type 2 diabetic nephropathy in Han population in Shanghai. J Shanghai Jiaotong Univ. 2008, 28: 376-379.
Wu LS, Hsieh CH, Pei D, Hung YJ, Kuo SW, Lin E: Association and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPARgamma and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetes. Nephrol Dial Transplant. 2009, 24: 3360-3366. 10.1093/ndt/gfp271.
De Cosmo S, Motterlini N, Prudente S, et al.: BENEDICT Study Group. Impact of the PPAR-gamma2 Pro12Ala polymorphism and ACE inhibitor therapy on new-onset microalbuminuria in type 2 diabetes: evidence from BENEDICT. Diabetes. 2009, 58: 2920-2929. 10.2337/db09-0407.
Liu L, Zheng T, Wang F, et al.: Pro12Ala polymorphism in the PPARG gene contributes to the development of diabetic nephropathy in Chinese type 2 diabetic patients. Diabetes Care. 2010, 33: 144-149. 10.2337/dc09-1258.
Lapice E, Pinelli M, Riccardi G, Vaccaro O: Pro12Ala polymorphism in the PPARG gene contributes to the development of diabetic nephropathy in Chinese type 2 diabetic patients: comment on the study by Liu et al. Diabetes Care. 2010, 33: e114-10.2337/dc10-0596.
Zhu W, Ji Y, Bu L, Yang Q: The correlativity study of peroxisome proliferator activated receptor g2 gene polymorphism with susceptibility of diabetic nephropathy in type 2 diabetes mellitus. J Radio-immunology. 2011, 24: 296-298.
De Cosmo S, Prudente S, Lamacchia O, et al.: PPAR{gamma}2 P12A polymorphism and albuminuria in patients with type 2 diabetes: a meta-analysis of case–control studies. Nephrol Dial Transplant. 2011, 26: 4011-4016. 10.1093/ndt/gfr187.
Zhang H, Zhu S, Chen J, Tang Y, Hu H, Mohan V, Venkatesan R, Wang J, Chen H: Peroxisome Proliferator–Activated Receptor g Polymorphism Pro12Ala Is Associated With Nephropathy in Type 2 Diabetes. Diabetes Care. 2012, 35: 1388-1393. 10.2337/dc11-2142.
Fujisawa T, Ikegami H, Kawaguchi Y, et al.: Meta-analysis of association of insertion/deletion polymorphism of angiotensin I-con-verting enzyme gene with diabetic nephropathy and retinopathy. Diabetologia. 1998, 41: 47-53. 10.1007/s001250050865.
Fei J, Li-Shuai Q, Xi-Zhong S: Association between the methylenetetrahydrofolate reductase C677T polymorphism and hepatocellular carcinoma risk: a meta-analysis. Diagn Pathol. 2009, 4: 39-10.1186/1746-1596-4-39.
de Matos L, Adriana Del G, Carolina M, de Lima Farah M, Auro Del G, da Silva Pinhal M: Expression of ck-19, galectin-3 and hbme-1 in the differentiation of thyroid lesions: systematic review and diagnostic meta-analysis. Diagn Pathol. 2012, 7: 97-10.1186/1746-1596-7-97.
Xue Q, Qiliu P, Aiping Q, Zhiping C, Liwen L, Yan D, Li X, Juanjuan X, Haiwei L, Taijie L, Shan L, Jinmin Z: Association of COMT Val158Met polymorphism and breast cancer risk: an updated meta-analysis. Diagn Pathol. 2012, 7: 136-10.1186/1746-1596-7-136.
Liwa Y, Jianqiu C: Association of MHTFR Ala222Val (rs1801133) polymorphism and breast cancer susceptibility: An update meta-analysis based on 51 research studies. Diagn Pathol. 2012, 7: 171-10.1186/1746-1596-7-171.
Acknowledgements
This work was not supported by any kind of fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
Both authors declared that they have no conflict interest in relation to this study.
Authors’ contributions
Lei Wang, Zan Teng, Shuang Cai,Difei Wang, Xin Zhao and Kai Yu drafted the manuscript, and carried out the molecular genetic studies, participated in the sequence alignment, Lei Wang reviewed the manuscript finally. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, L., Teng, Z., Cai, S. et al. The association between the PPARγ2 Pro12Ala polymorphism and nephropathy susceptibility in type 2 diabetes: a meta-analysis based on 9,176 subjects. Diagn Pathol 8, 118 (2013). https://doi.org/10.1186/1746-1596-8-118
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-1596-8-118