Abstract
Background
Sirtuin1 (SIRT1) regulates gene expression in distinct metabolic pathways and mediates beneficial effects of caloric restriction in animal models. In humans, SIRT1 genetic variants associate with fasting energy expenditure. To investigate the relevance of SIRT1 for human metabolism and caloric restriction, we analyzed SIRT1 genetic variants in respect to the outcome of a controlled lifestyle intervention in Caucasians at risk for type 2 diabetes.
Methods
A total of 1013 non-diabetic Caucasians from the Tuebingen Family Study (TUEF) were genotyped for four tagging SIRT1 SNPs (rs730821, rs12413112, rs7069102, rs2273773) for cross-sectional association analyses with prediabetic traits. SNPs that associated with basal energy expenditure in the TUEF cohort were additionally analyzed in 196 individuals who underwent a controlled lifestyle intervention (Tuebingen Lifestyle Intervention Program; TULIP). Multivariate regressions analyses with adjustment for relevant covariates were performed to detect associations of SIRT1 variants with the changes in anthropometrics, weight, body fat or metabolic characteristics (blood glucose, insulin sensitivity, insulin secretion and liver fat, measured by magnetic resonance techniques) after the 9-month follow-up test in the TULIP study.
Results
Minor allele (X/A) carriers of rs12413112 (G/A) had a significantly lower basal energy expenditure (p = 0.04) and an increased respiratory quotient (p = 0.02). This group (rs12413112: X/A) was resistant against lifestyle-induced improvement of fasting plasma glucose (GG: -2.01%, X/A: 0.53%; p = 0.04), had less increase in insulin sensitivity (GG: 17.3%, X/A: 9.6%; p = 0.05) and an attenuated decline in liver fat (GG: -38.4%, X/A: -7.5%; p = 0.01).
Conclusion
SIRT1 plays a role for the individual lifestyle intervention response, possibly owing to decreased basal energy expenditure and a lower lipid-oxidation rate in rs12413112 X/A allele carriers. SIRT1 genetic variants may, therefore, represent a relevant determinant for the response rate of individuals undergoing caloric restriction and increased physical activity.
Similar content being viewed by others
Background
The sirtuin SIRT1 (silent mating type information regulation 2 homolog 1) is a NAD+-dependent deacetylase involved in regulation of glucose metabolism in different organs [1, 2]. SIRT1 directs its deacetylase activity against histones, a process that facilitates the formation of heterochromatin with subsequent repression of gene transcription. In addition, SIRT1 directly regulates activity of metabolic transcription factors and their co-activators, as e. g. forkhead transcription factor FOXO1, nuclear factor NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B-cells 1) or PGC-1α (peroxisome proliferator-activated receptor gamma coactivator 1 alpha).
Due to its ubiquitous expression, SIRT1 activity is relevant for many insulin-sensitive organs. In adipose tissue, SIRT1 down-regulates fat storage by increased lipolysis via the inactivation of PPARγ (peroxisome proliferator-activated receptor gamma) [3]. In pancreatic β-cells, SIRT1 enhances glucose-stimulated insulin secretion via down-regulation of UCP2 (uncoupling protein 2) [4]. In the liver, particularly under low-nutrient conditions, SIRT1 induces gluconeogenesis and inhibits glycolysis by deacetylating PGC-1α [5]. In skeletal muscle of obese mice, SIRT1 mediates the insulin sensitizing effect of resveratrol. The antioxidant resveratrol induces increased exercise endurance and higher basal energy expenditure in mice, thereby promoting resistance against diet-induced obesity [6] and mortality [7]. Interestingly, modest overexpression of SIRT1 also protects mice against high-fat diet induced glucose intolerance and hepatic steatosis [8], so that SIRT1 may represent a promising future pharmacological target to prevent the metabolic sequelae of chronic exposure to a high-fat diet.
Summarizing all these data, one may assume that human SIRT1 genetic variants may also play a role in lifestyle intervention response in humans. So far, no studies exist on whether SIRT1 genetic variants affect the individual metabolic susceptibility to caloric restriction or increased physical activity. The only genetic study that assessed SIRT1 genetic variants in the context of metabolism and diabetes reports an association of SIRT1 genetics with basal energy expenditure in Finnish type 2 diabetic patients [6]. We, therefore, conducted the present study to investigate SIRT1 variants in the context of a controlled lifestyle intervention in prediabetic individuals at high risk for later onset of diabetes. For this purpose, 1013 subjects from the cross-sectional Tuebingen Family Study (TUEF) were genotyped for SIRT1 tagging SNPs and association analyses for energy expenditure and other metabolic traits were undertaken. In a second step, genotypes that associated with different phenotypes in the TUEF cohort were analyzed in the ongoing Tübingen Lifestyle Intervention Programme (TULIP) [9] for longitudinal changes of blood glucose, insulin secretion, insulin sensitivity and liver fat.
Methods
Study design and participants
All volunteers signed written consent to the study protocol, approved by the local medical ethics committee. Genotyping for SIRT1 was done in 1013 study participants, 96 of which were excluded (manifest diabetes or missing data), resulting in a total cohort of n = 917 for cross-sectional analyses. 196 participants of the study cohort also participated in TULIP, so that longitudinal (9 month) data on metabolic traits including liver fat content were available. TULIP participants are at increased risk of type 2 diabetes (family history of type 2 diabetes, body mass index (BMI) > 27 kg/m2, diagnosis of IGT or previous diagnosis of gestational diabetes) and undergo a controlled lifestyle intervention designed for the same targets as the Diabetes Prevention Study [10]: body weight reduction by > 5%, dietary fat (saturated fatty acids) reduction to < 30% (< 10%) of caloric intake, increase of dietary fibre (> 15 g/1000 kcal) and enhanced physical activity (> 3 h/week; see [9] for further details on TULIP).
Oral glucose tolerance test (OGTT)
All participants underwent an OGTT performed according to WHO recommendations [11]. Blood glucose, plasma insulin, plasma C-peptide and non-esterified fatty acid (NEFA) levels were determined at 0, 30, 60, 90 and 120 min intervals.
Insulin sensitivity, insulin secretion, and analytical methods
Blood glucose was determined using a bedside glucose analyzer (Yellow Springs Instruments, Yellow Springs, CO, USA). Plasma insulin was determined by microparticle enzyme immunoassay (Abbott Laboratories, Tokyo, Japan), and plasma C-peptide was determined by radioimmunoassay (Byk-Sangtec, Dietzenbach, Germany). Insulin sensitivity was estimated using the model from Matsuda and DeFronzo [12], and insulin secretion was estimated from 30 min C-peptide levels obtained during the OGTT. Basal energy expenditure and respiratory quotient were measured with a DELTATRAC™ Metabolic Monitor (Hoyer; Bremen, Germany) under resting conditions. Further details on analytical procedures are provided elsewhere [13].
Determination of body and liver fat
Total body fat and lean body mass were measured by bioelectrical impedance (RJL; Detroit, MI, USA). Liver fat was determined by magnetic resonance spectroscopy (1.5T Magnetom Sonata; Siemens, Erlangen, Germany) in the posterior 7th segment of the liver. A single-voxel stimulated echo acquisition mode technique was applied (repetition time = 4 s, echo time = 10 msec, 32 acquisitions) for a voxel of 3 × 3 × 2 cm3, and liver fat content was calculated by the signal integral (methylene/methyl signals between 0.7–1.5 ppm) in reference to the sum of water and lipid signal integrals [14].
Genotyping
The TaqMan assay (Eurogentec; Liege, Belgium) and the fluorescence detecting ABI Prism 7500 (Applied Biosystems; Foster City, CA, USA) were used for genotyping of SNPs selected by HapMap analysis (phase I, release 21a, January 2007). A minor allele frequency (MAF) > 0.05 and a linkage disequilibrium measure (r2) < 0.8 were prerequisites for tagging SNP selection in a region comprising the whole SIRT1 gene and 10 kb of its promoter. To monitor TaqMan test reproducibility and accuracy, samples of 50 individuals were sequenced for the selected tagging SNPs with an ABI Prism 310 genetic analyser (Applied Biosystems). Subsequently, one sample per allelic combination was included in each TaqMan assay as a known reference genotype control.
Statistics
Data are presented as means ± SD. The Hardy-Weinberg equilibrium was calculated by the χ2-test. Multivariate linear regression analyses with adjustments for relevant covariates were undertaken, using logarithmically transformed data for non-normally distributed parameters. The JMP 4.0.4 software (SAS Institute; Cary, NC, USA) was used for statistical analyses, and p values ≤ 0.05 were considered statistically significant.
Results
SIRT1genetic variants in the TUEF/TULIP cohort
Four SNPS of the SIRT1 gene (~35 kb, 9 exons) on chromosome 10q21.3 were selected from the HapMap according the selection criteria for genotyping: rs730821 (A/G, MAF = 0.186 in the TUEF cohort, promoter), rs12413112 (G/A, MAF = 0.114, intron4), rs7069102 (G/C, MAF = 0.301, intron4) and rs2273773 (T/C, MAF = 0.058, intron5). All tagging SNPs were in Hardy-Weinberg-Equilibrium (p > 0.21; all, both TUEF and TULIP cohort), and linkage disequilibrium (r2) ranged from 0.008 to 0.53.
Cross-sectional TUEF study
We first verified whether SIRT1 genetic variants associate with basal energy expenditure in the TUEF cohort, as shown before in a Finnish population [6]. Indeed, basal energy expenditure was significantly lower in rs12413112 (p = 0.04; dominant model) and rs7069102 (p = 0.05; dominant model) minor allele carriers compared to homozygous carriers of the major G alleles, after adjustment for relevant covariates as age, sex, lean body mass and body fat [15–17]. Minor allele carriers of rs12413112, in addition, had a significantly increased respiratory quotient (RQ; +2.2%, p = 0.02; dominant model; see Table 1). In contrast, anthropometrics (BMI, body fat, waist-to-hip ratio) did not differ among SIRT1 genotypes. There was no significant association with (fasting and 2 h OGTT) glucose and insulin plasma levels, insulin sensitivity (OGTT and clamped) or insulin secretion (dominant p for all > 0.12; Table 1). Serum lipid parameters (total-, HDL-, LDL-cholesterol, apolipoproteins, NEFAs) did also not differ between SIRT1 genotypes (data not shown).
SIRT1variants and TULIP lifestyle intervention outcome traits
The two SIRT1 SNPs that associated with energy expenditure, namely rs12413112 and rs7069102, were subsequently analyzed in 196 individuals (118 females, 78 males, aged 45.8 ± 11.2 years; GG: n = 151, GA: n = 42, AA: n = 3 individuals) from the TULIP study cohort who had completed the 9-month follow-up test. In this cohort, average weight loss amounted to 2.56 ± 4.06 kg in a follow-up time of 259 ± 53 days, resulting in a BMI reduction of 0.87 ± 1.38.
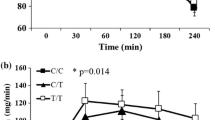
We did not detect significant associations between rs12413112 genotype and weight (p = 0.37) or BMI (p = 0.38) change. By contrast, minor allele carriers of rs12413112 were resistant against lifestyle-induced improvement of fasting plasma glucose (GG: -2.01%, X/A: 0.53%; p = 0.04, adjusted for age, sex, follow-up time, BMI at baseline and follow-up and fasting plasma glucose at baseline; Fig. 1A), had a weaker increase in insulin sensitivity (GG: 17.3%, X/A: 9.6%; p = 0.05, adjusted for age, sex, follow-up time, BMI at baseline and follow-up and insulin sensitivity at baseline; Fig. 1B) and a remarkably attenuated decline in liver fat content (GG: -38.4%, X/A: -7.5%; p = 0.01, adjusted for age, sex, follow-up time, BMI at baseline and follow-up and liver fat content at baseline; Fig. 1C). The effect on liver fat content remained significant even after Bonferroni's correction for multiple testing (2 tests: energy expenditure and obesity), and we had a power of 0.76 to detect this effect. The rs12413112 genotype effects on fasting glucose (0.50) and insulin sensitivity (0.56) change did not show a sufficient statistical power and also did not remain significant after correction for multiple comparisons. The rs7069102 genotype did not significantly associate with longitudinal changes in weight (p = 0.40; dominant model), BMI (p = 0.41), fasting glucose (p = 0.08), insulin sensitivity (p = 0.45) and liver fat (p = 0.09) in the longitudinal TULIP analysis.
Interaction of rs12413112 SIRT1 genotype with the metabolic responses during a controlled 9-month lifestyle intervention. Rs12413112 SIRT1 genotype (GG): filled circles (●); X/A: opened circles (○). a) Response of plasma glucose determined during the fasting state. b) Response of insulin sensitivity estimated by the formula from Matsuda et al. [12]. c) Response of liver fat content, determined by magnetic resonance spectroscopy. For statistical analyses, all data were log-transformed and adjusted for age, sex, follow-up-time, BMI at both baseline and follow-up and the corresponding variable (plasma glucose, insulin sensitivity or liver fat) measured at baseline. Data are presented as means+SEM (G/G: n = 152, X/A: n = 45 TULIP participants).
Discussion
This is the first study to have analysed the impact of SIRT1 genetic variants on the outcome of a lifestyle intervention in a diabetes risk population. Despite numerous works demonstrating SIRT1 function in different insulin-responsive tissues [1, 3–5, 18, 19], we were not able to verify a significant association of SIRT1 variants with plasma glucose, plasma insulin, NEFAs, serum lipids, insulin sensitivity or insulin secretion, both under fasting conditions and 2 h OGTT in the cross-sectional analyses. However, minor allele carriers of rs12413112 had significantly lower basal energy expenditure and an increased respiratory quotient (RQ) at baseline. Interestingly, in a Finnish population, higher basal energy expenditure was reported for three SIRT1 SNPs [6]. This difference may be due to the very low linkage disequilibrium of these three SNPs with rs12413112, ranging from 0.0076 to 0.10 in both the TUEF cohort as well as the CEU population of the HapMap project, so that rs12413112 may act in the opposing direction on SIRT1 function compared to the previously reported SNPs. In addition, our study cohort has considerably differing allele frequencies compared to the investigated Finnish population, at least in the SIRT1 locus.
The minor allele of rs12413112 is associated with unresponsiveness of fasting plasma glucose, insulin sensitivity and liver fat to a controlled 9 month lifestyle intervention. Because of the higher RQ in this subgroup, one may speculate that the rs12413112 minor A-allele contributes to a lower rate of lipid-oxidation, resulting in the observed lower liver fat reduction, despite dietary changes and increased physical activity. Taking into account that modest overexpression of SIRT1 leads to protection from high-fat diet induced hepatic steatosis in mice [8], our study let assume that the minor allele of rs12413112 may negatively affect SIRT1 expression, especially as the SNPs tagged by this intronic SNP, namely rs11599151, rs11599524 and rs12778366, are all located in the SIRT1 promoter.
Finally, the rather small study sample size has to be mentioned as the major statistical limitation of this study. The here reported effects of SIRT1 variants are statistically significant, but a conservative adjustment for multiple comparisons would render the associations in TUEF with energy expenditure and RQ non-significant. However, we think that our findings are relevant, as the TULIP study provides a very precisely phenotyped and closely monitored lifestyle intervention cohort which is suitable for studying the role of genetic variants by a classical candidate-gene approach.
Conclusion
In summary, our study shows that SIRT1 genetic variants may affect individual susceptibility to caloric restriction and increased physical activity. Although the cross-sectional study let rather doubt that SIRT1 does represent a classical diabetes risk gene, SIRT1 genetic variants may determine the individual response to a lifestyle intervention. Beyond AdipoR1 [14] and PPARδ [20], SIRT1 is the third gene detected which does not affect prediabetic traits in a cross-sectional study approach, but which remarkably influences the response to a lifestyle intervention. Therefore, SIRT1 should be elucidated in other prospective lifestyle intervention studies, and further research on SIRT1 as a potential pharmacological target for prevention of type 2 diabetes is warranted.
References
Yang T, Fu M, Pestell R, Sauve AA: SIRT1 and endocrine signaling. Trends Endocrinol Metab. 2006, 17 (5): 186-191. 10.1016/j.tem.2006.04.002.
Yamamoto H, Schoonjans K, Auwerx J: Sirtuin functions in health and disease. Mol Endocrinol. 2007, 21 (8): 1745-1755. 10.1210/me.2007-0079.
Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L: Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004, 429 (6993): 771-776. 10.1038/nature02583.
Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L: Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006, 4 (2): e31-10.1371/journal.pbio.0040031.
Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P: Nutrient control of glucose homeostasis through a complex of PGC- 1alpha and SIRT1. Nature. 2005, 434 (7029): 113-118. 10.1038/nature03354.
Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J: Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006, 127 (6): 1109-1122. 10.1016/j.cell.2006.11.013.
Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA: Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006, 444 (7117): 337-342. 10.1038/nature05354.
Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH: Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008, 105 (28): 9793-9798. 10.1073/pnas.0802917105.
Schafer S, Kantartzis K, Machann J, Venter C, Niess A, Schick F, Machicao F, Haring HU, Fritsche A, Stefan N: Lifestyle intervention in individuals with normal versus impaired glucose tolerance. European Journal of Clinical Investigation. 2007, 37 (7): 535-543. 10.1111/j.1365-2362.2007.01820.x.
Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa M: Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001, 344 (18): 1343-1350. 10.1056/NEJM200105033441801.
Alberti KG, Zimmet PZ: Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998, 15 (7): 539-553. 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S.
Matsuda M, DeFronzo RA: Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999, 22 (9): 1462-1470. 10.2337/diacare.22.9.1462.
Weyrich P, Machicao F, Staiger H, Simon P, Thamer C, Machann J, Schick F, Guirguis A, Fritsche A, Stefan N, Haring HU: Role of AMP-activated protein kinase gamma 3 genetic variability in glucose and lipid metabolism in non-diabetic whites. Diabetologia. 2007, 50 (10): 2097-2106. 10.1007/s00125-007-0788-8.
Stefan N, Machicao F, Staiger H, Machann J, Schick F, Tschritter O, Spieth C, Weigert C, Fritsche A, Stumvoll M, Haring HU: Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia. 2005, 48 (11): 2282-2291. 10.1007/s00125-005-1948-3.
Ravussin E, Lillioja S, Anderson TE, Christin L, Bogardus C: Determinants of 24-hour energy expenditure in man. Methods and results using a respiratory chamber. J Clin Invest. 1986, 78 (6): 1568-1578. 10.1172/JCI112749.
Luke A, Durazo-Arvizu R, Cao G, Adeyemo A, Tayo B, Cooper R: Positive association between resting energy expenditure and weight gain in a lean adult population. Am J Clin Nutr. 2006, 83 (5): 1076-1081.
Weyer C, Snitker S, Rising R, Bogardus C, Ravussin E: Determinants of energy expenditure and fuel utilization in man: effects of body composition, age, sex, ethnicity and glucose tolerance in 916 subjects. Int J Obes Relat Metab Disord. 1999, 23 (7): 715-722. 10.1038/sj.ijo.0800910.
Zhang J: The Direct Involvement of SirT1 in Insulin-induced Insulin Receptor Substrate-2 Tyrosine Phosphorylation. J Biol Chem. 2007, 282 (47): 34356-34364. 10.1074/jbc.M706644200.
Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q: SIRT1 Improves Insulin Sensitivity under Insulin-Resistant Conditions by Repressing PTP1B. Cell Metab. 2007, 6 (4): 307-319. 10.1016/j.cmet.2007.08.014.
Thamer C, Machann J, Stefan N, Schafer SA, Machicao F, Staiger H, Laakso M, Bottcher M, Claussen C, Schick F, Fritsche A, Haring HU: Variations in PPARD determine the change in body composition during lifestyle intervention: a whole-body magnetic resonance study. J Clin Endocrinol Metab. 2008, 93 (4): 1497-1500. 10.1210/jc.2007-1209.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/9/100/prepub
Acknowledgements
We thank all the research volunteers for their participation. We gratefully acknowledge the technical assistance of Melanie Weisser, Alke Guirguis, Roman-Georg Werner and Anna Bury. Norbert Stefan is currently supported by a Heisenberg Grant of the Deutsche Forschungsgemeinschaft (DFG). The TULIP study is supported by a grant from the DFG (KFO 114 and Heisenberg-Grant to Norbert Stefan 1096/1-1) and the European Community's FP6 EUGENE6 (LSHM-CT-2004-512013). The authors would like to thank all participants for their cooperation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PW was responsible for the complete statistical analysis and prepared all tables, figures and the manuscript. FM is responsible of the genotyping facility at the University of Tübingen. JR made contributions to phenotyping of TULIP participants. JM and FS performed the magnetic resonance spectroscopy studies for liver fat determination. OT contributed to the statistical analysis. NS, AF and HUH acted as principal investigators for the TUEF and TULIP study, and were responsible for patient and data management. All authors contributed to the preparation of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Weyrich, P., Machicao, F., Reinhardt, J. et al. SIRT1 genetic variants associate with the metabolic response of Caucasians to a controlled lifestyle intervention – the TULIP Study. BMC Med Genet 9, 100 (2008). https://doi.org/10.1186/1471-2350-9-100
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-9-100