Abstract
Aims/hypothesis
The adipokine adiponectin has insulin-sensitising, anti-atherogenic and anti-inflammatory properties. Recently, the genes for mouse and human adiponectin receptor-1 (ADIPOR1) and -2 (ADIPOR2) have been cloned. The aim of this study was to investigate whether genetic variants of the genes encoding ADIPOR1 and ADIPOR2 play a role in human metabolism.
Materials and methods
We screened ADIPOR1 and ADIPOR2 for polymorphisms and determined their association with glucose metabolism, lipid metabolism, an atherogenic lipid profile and inflammatory markers in 502 non-diabetic subjects. A subgroup participated in a longitudinal study; these subjects received diet counselling and increased their physical activity.
Results
We identified six variants of ADIPOR1 and seven variants of ADIPOR2. A single-nucleotide polymorphism (SNP) in the putative promoter region 8503 bp upstream of the translational start codon (−8503 G/A) of ADIPOR1 (frequency of allele A=0.31) was in almost complete linkage disequilibrium with another SNP (−1927 T/C) in intron 1. Subjects carrying the −8503 A and −1927 C alleles had lower insulin sensitivity, as estimated from a 75 g OGTT (p=0.04) and determined during a euglycaemic clamp (n=295, p=0.04); they also had higher HbA1c levels (p=0.02) and, although the difference was not statistically significant, higher liver fat (n=85, determined by proton magnetic resonance spectroscopy, p=0.056) (all p values are adjusted for age, sex and percentage of body fat). In the longitudinal study (n=45), the −8503 A and −1927 C alleles were associated with lower insulin sensitivity (p=0.03) and higher liver fat (p=0.02) at follow-up compared with the −8503 G and −1927 T alleles, independently of basal measurements, sex and baseline and follow-up percentage of body fat.
Conclusions/interpretation
The present findings suggest that the −8503 G/A SNP in the promoter or the −1927 T/C SNP in intron 1 of ADIPOR1 may affect insulin sensitivity and liver fat in humans.
Similar content being viewed by others
Introduction
Adiponectin is an adipokine that regulates glucose metabolism, lipid metabolism and inflammation. Adiponectin knock-out mice exhibit severe diet-induced insulin resistance [1, 2], and administration of recombinant adiponectin to both lipoatrophic and obese rodents is followed by improvements in insulin sensitivity and glycaemia [2–6]. In addition, adiponectin was shown to inhibit inflammatory pathways and to reduce atherosclerosis in rodents [7, 8]. Low plasma adiponectin concentrations are associated with insulin resistance in cross-sectional analyses in healthy humans [7, 8] and predict an increase in insulin resistance [9, 10] and type 2 diabetes [11, 12]. In addition, plasma adiponectin concentrations are negatively associated with TNF-α and C-reactive protein levels, and positively with plasma HDL cholesterol concentrations [7, 8].
While the role of adiponectin in animal metabolism and associations of adiponectin levels with glucose and lipid metabolism in humans have been studied extensively, information on adiponectin receptors was not available until June 2003. The genes for the mouse and human adiponectin receptor-1 (ADIPOR1) and-2 (ADIPOR2) have been cloned [13]. Both receptors mediate effects of globular and/or full-length adiponectin. In the mouse, mRNA expression of ADIPOR1 was predominantly found in skeletal muscle. The highest concentrations of ADIPOR2 mRNA were found in the liver. In contrast, in human tissues the highest mRNA levels of both receptors were found in skeletal muscle [13]. These findings were confirmed in a recent study [14]. In addition, the expression levels of both receptors were found to correlate positively with insulin sensitivity in that study. This suggests that adiponectin receptors may also modulate insulin sensitivity in humans. However, insulin was shown to affect expression of adiponectin receptors in insulin target organs, such as skeletal muscle and liver. Insulin deficiency increased and insulin replenishment reduced the expression of ADIPOR1 and ADIPOR2 in animals in vivo, involving the insulin/phosphoinositide 3-kinase/Foxo1 pathway [15]. Thus, the question of whether the association of the expression level of adiponectin receptors with insulin sensitivity is merely a result of insulinaemia is still under investigation.
One approach to determining the role of adiponectin receptors in human metabolism is to investigate whether polymorphisms in ADIPOR1 and ADIPOR2 exist and to test whether these polymorphisms are associated with features of the metabolic syndrome, particularly, with insulin resistance. Intramyocellular and intrahepatic lipids were found to be important determinants of insulin resistance in humans [16–18]. Adiponectin was shown to increase lipid oxidation in these tissues in animals and in vitro. Therefore, we further investigated whether polymorphisms in ADIPOR1 and ADIPOR2 are associated with intramyocellular and intrahepatic lipid, as determined by proton nuclear magnetic resonance spectroscopy. First, we determined the relationship of the phenotypes with polymorphisms in ADIPOR1 and ADIPOR2 in a cross-sectional study. Secondly, we investigated whether the polymorphisms were associated with changes in phenotype in a longitudinal study that included subjects who were on an intervention to reduce adiposity and to prevent type 2 diabetes.
Subjects and methods
Subjects
In the cross-sectional study we analysed data from a total of 502 people of European extraction who had NGT or IGT. The individuals were recruited from the southern part of Germany and participated in the ongoing Tübingen Family Study for type 2 diabetes. The participants were not taking any medication known to affect glucose tolerance or insulin sensitivity. They were considered healthy according to a physical examination and routine laboratory tests. All subjects first underwent an OGTT. When they were found not to have diabetes they underwent further tests. Using self-reported data, only 1.2% of the subjects were related and 72% had a family history of type 2 diabetes. A subgroup of 295 individuals had measurements of insulin sensitivity obtained during a euglycaemic–hyperinsulinaemic clamp.
In the longitudinal study, a group of 45 subjects underwent dietary counselling and increased their physical activity to decrease adiposity and to prevent type 2 diabetes. Data from these individuals were also included in the cross-sectional study. The individuals had a family history of type 2 diabetes and/or a BMI greater than 27 kg/m2. These subjects underwent baseline measurements and had regular visits to our clinic thereafter. The dietary counselling aimed to reduce the total intake of calories, particularly the intake of calories from fat. Subjects were instructed to increase physical activity and to perform at least 3 h of sports per week. The participants did not take any medication known to affect glucose tolerance or insulin sensitivity. Tests were performed at 07.00 h after an overnight fast of 12 h. Informed written consent was obtained from all participants and the local medical ethics committee had approved the studies.
Body composition and body fat distribution
Body composition was measured by bioelectrical impedance as the percentage of body fat. BMI was calculated as weight divided by the square of height (kg/m2). Waist and hip circumferences were measured in the supine position, and WHR was calculated as an index of body fat distribution.
OGTT
All subjects underwent a 75 g OGTT and venous blood samples were obtained at 0, 30, 60, 90 and 120 min for determination of plasma glucose, insulin and C-peptide. Glucose tolerance was determined according to the 1997 World Health Organization diagnostic criteria [19]. Insulin sensitivity was calculated from glucose and insulin values during the OGTT, as proposed by Matsuda and DeFronzo [20]. The 30-min C-peptide plasma concentrations during the OGTT and the first phase of insulin secretion determined from the OGTT (1283+1.829 Ins30−138.7 Glu30+3.772 Ins0), as described earlier [21], were used as an estimate of beta-cell function.
Euglycaemic–hyperinsulinaemic clamp
After a 12-h overnight fast, an antecubital vein was cannulated for infusion of insulin and glucose. A dorsal hand vein of the contralateral arm was cannulated and placed under a heating device to permit sampling of arterialised blood. Subjects received a primed insulin infusion at the rate of 1.0 mU kg−1 min−1 for 2 h. Blood was drawn every 5 min for determination of blood glucose, and a glucose infusion was adjusted appropriately to maintain the fasting glucose level. An insulin sensitivity index (ISI; in μmol kg−1 min−1 [pmol/l]−1) for systemic glucose uptake was calculated as the mean infusion rate of glucose (in μmol kg−1 min−1) necessary to maintain euglycaemia during the last 60 min of the euglycaemic–hyperinsulinaemic clamp divided by the steady-state plasma insulin concentration.
Intrahepatic and intramyocellular lipid content
Liver fat was determined by localised proton magnetic resonance spectroscopy using a 1.5 T whole-body imager (Magnetom Sonata, Siemens Medical Solutions, Erlangen, Germany). For volume selection, a single-voxel stimulated echo acquisition mode (STEAM) technique was applied (repetition time [TR]=4 s, echo time [TE]=10 ms, 32 acquisitions) and a voxel of 3×3×2 cm3 was placed in the posterior part of the seventh segment of the liver. Subjects were asked to breathe within the TR interval and to be in expiration during data acquisition. The liver fat was assessed quantitatively by analysing the signal integrals of methylene and methyl resonances (between 0.7 and 1.5 ppm), using the liver water signal integral at 4.8 ppm as internal reference.
Intramyocellular lipid content (IMCL) and lipid content interlaced between the muscle fibres (extramyocellular lipid content, EMCL) was differentiated because of their geometrical arrangement using proton magnetic resonance spectroscopy as previously described [22]. In brief, localised image-guided proton spectra of the tibialis anterior muscle representing a muscle of mixed type I and II fibres and of the soleus muscle representing a muscle of predominantly type I fibres with high oxidative capacity were acquired on a 1.5 T whole-body imager (Magnetom Vision; Siemens Medical Solutions, Erlangen, Germany). For volume selection, a single-voxel STEAM technique was applied. Measurement parameters were TE=10 ms, TR=2 s, volume of interest 11×11×20 mm3, 40 acquisitions. IMCL and EMCL were quantified on the separation of two resonances in the lipid–CH2-region.
Analytical procedures
Blood glucose was determined using a bedside glucose analyser (glucose-oxidase method; Yellow Springs Instruments, Yellow Springs, CO, USA). Plasma insulin was determined by microparticle enzyme immunoassay (Abbott Laboratories, Tokyo, Japan) and plasma C-peptide by radioimmunoassay (Byk-Sangtec, Dietzenbach, Germany). Serum samples were frozen immediately and stored at −20°C for determination of adiponectin by radioimmunoassay (LINCO Research, St Charles, MO, USA) and free fatty acids with an enzymatic method (Wako Chemicals, Neuss, Germany). Plasma TNF-α and IL-6 concentrations were determined by enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, MN, USA).
Screening of ADIPOR1/ADIPOR2 and genotyping of single-nucleotide polymorphisms
Human ADIPOR1 is composed of eight exons spanning 17.5 kb on chromosome 1p36.13-q41. Human ADIPOR2 maps to chromosome 12p13.31, comprises eight exons and spans 97.6 kb. Sequencing was done according to the sequences NT_004671.15 (ADIPOR1) and NT_009759.15 (ADIPOR2) in the NCBI database (http://www.ncbi.nih.gov/entrez/query.fcgi?db=gene, last accessed in August 2005). We performed 14 PCR amplifications of ADIPOR1 and 13 of ADIPOR2 with a series of specific primer pairs using DNA extracted from blood leucocytes in 50 subjects. For the two genes, primer sequences cover 2 kb of the proximal promoter, all exons, exon–intron boundaries (100–200 bp of the flanking intronic region), including the 5′ untranslated region and part of the 3′ untranslated region. The sequence of ADIPOR1 in the NCBI database was updated towards the end of 2004, resulting in reorganisation of the exon order. Since we started our sequencing before this, we initially considered a large region upstream of exon 2 as a promoter and, therefore, additionally sequenced intron 1.
The polymorphisms were localised by direct sequencing. PCR products were sequenced bidirectionally, using an ABI Prism dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA, USA), and analysed on an automated sequencer (ABI model 310). Polymorphisms in selected DNA samples were genotyped using the TaqMan assay (Applied Biosystems). The TaqMan genotyping reaction was amplified on a GeneAmp PCR system 7000 (50°C for 10 min, 95°C for 10 min, 92°C for 15 s, 60°C for 1 min for 35 cycles) and fluorescence was detected on an ABI Prism 7000 sequence detector (Applied Biosystems). As a quality standard, we randomly included six positive and two negative (all components excluding DNA) sequenced controls in each TaqMan reader assay. All controls were correctly identified.
Statistical analyses
Unless otherwise stated, data are given as mean±SE. Except for insulin secretion (analysed only in individuals with normal glucose tolerance [NGT]), subjects with NGT and impaired glucose tolerance (IGT) were analysed together. Hardy–Weinberg equilibrium was tested with the χ 2 test. Statistical comparison was performed using logarithmically transformed data (for non-normally distributed parameters). Differences in anthropometrics and metabolic characteristics between genotypes were tested using general linear regression models. In these models the trait was the dependent variable, whereas age, sex and genotype, for example, were the independent variables. An additive model was used in all cross-sectional analyses for all SNPs. In the longitudinal study, differences between measurements at baseline and follow-up were tested with the two-tailed Student’s t-test. Because of the small number of subjects who had follow-up data, subjects who were homozygous for the mutant alleles (e.g. n=3 for the −8503 G/A polymorphism) and heterozygotes were combined for analyses. In general linear regression models, insulin sensitivity and liver fat at follow-up were the dependent variables and insulin sensitivity and liver fat at baseline, sex and genotype were the independent variables. Pairwise linkage disequilibrium (D′, r 2) and haplotypes were determined using the THESIAS program [23] and the Java linkage disequilibrium plotter (http://www.genepi.com.au/projects/jlin, last accessed in August 2005). THESIAS performs haplotype-based association analyses in unrelated individuals with phase-unknown genotypes. It is based on a maximum likelihood model and is linked to the SEM algorithm. This allows the simultaneous estimation of haplotype frequencies and of their associated effects on the phenotype of interest. Both quantitative and qualitative phenotypes can be studied. Using this approach the differences in the phenotypes of the most common (frequency >0.02) haplotypes in respect to the so-called wild-type haplotype (this haplotype consisted only of the major alleles) was determined.
Because of the relatively small number of subjects in the longitudinal study, haplotype analyses were performed only in the cross-sectional study. A p value below 0.05 was considered statistical significant. The statistical software package JMP (SAS Institute, Cary, NC, USA) or SPSS version 10.0 software (SPSS, Chicago, IL, USA) was used.
Results
Screening for polymorphisms and genotyping results
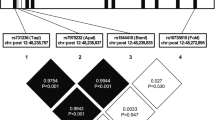
We identified six SNPs in ADIPOR1 (Table 1; Fig. 1a). Among them, three SNPs were already reported in the dbSNP database (http://ncbi.nlm.nih.gov/projects/SNP, last accessed in August 2005) and one SNP (9512 G/A) was recently found by Wang and colleagues [24] (Table 1). Pairwise linkage disequilibrium for the SNPs is shown in Table 2. The −8503 G/A SNP in the promoter region was in almost complete linkage disequilibrium with one novel SNP in intron 1 (−1927 T/C, frequency of allele C=0.31). All SNPs in ADIPOR1 were in Hardy–Weinberg equilibrium (p>0.77).
Schematic illustrations of ADIPOR1 (a) and ADIPOR2 (b) indicating the genetic variants detected in the genes and flanking regions. Grey boxes, exons; black boxes, sequence translated into the protein. Introns and flanking regions are shown as lines connecting the boxes. The SNPs are labelled with their position relative to the ATG start codon for those in the promoter, the introns and the 3′ untranslated region, with their position relative to the coding amino acid in the exons, with dbSNP database number, if applicable (http://www.ncbi.nlm.nih.gov/projects/SNP, last accessed in August 2005) or are indicated as ‘novel’. The allelic frequencies of the minor alleles are shown at the bottom of each panel
Seven SNPs were identified in ADIPOR2 (Table 1; Fig. 1b). Among them, three SNPs were already reported in the dbSNP database. The other SNPs are novel (Table 1). Pairwise linkage disequilibrium of the SNPs is shown in Table 2. The SNPs found in ADIPOR2 were typed after the typing of the SNPs in ADIPOR1 was completed. Because of technical problems, the DNA samples of 23 subjects were lost, and only 479 individuals could be typed for six SNPs in ADIPOR2. The SNP 29561 A/T in exon 7 was not typed because of its low allelic frequency (<0.0004). The SNPs in ADIPOR2 were also in Hardy–Weinberg equilibrium (p>0.32).
Cross-sectional study
A total of 502 subjects were included in the cross-sectional analyses. The population consisted of 209 males and 293 females with a mean age of 36±1 years (mean±SE) and a mean BMI of 26.4±0.3 kg/m2. Of the 502 subjects, 446 had NGT and 56 had IGT.
In ADIPOR1 the A allele at SNP −8503 G/A in the promoter and the C allele at SNP −1927 T/C in intron 1, but none of the alleles at the other SNPs were associated with lower insulin sensitivity estimated from the OGTT (p=0.04) and determined during the clamp (p=0.04; Table 3 and Electronic supplementary material [ESM] Fig. 1a,b). These alleles were also associated with higher plasma HbA1c (p=0.02; Table 3 and ESM Fig. 1c).
In 85 subjects liver fat was positively associated with percentage of body fat before (r=0.30, p=0.005) and after adjustment for age and sex (r=0.39, p=0.0002). It was negatively associated with insulin sensitivity (clamp) before (r=−0.62, p<0.0001) and after adjustment for age, sex and percentage of body fat (r=−0.56, p<0.0001). Although not statistically significant, the A allele at SNP −8503 G/A and the C allele at SNP −1927 T/C were also associated with higher liver fat (−8503 G/A: 1.23±0.14% [G/G], 1.22±0.16% [G/A] and 2.16±0.36% [A/A], log, p=0.056) (Fig. 2) after adjustment for age, sex and percentage of body fat.
Association of liver fat, determined by proton magnetic resonance spectroscopy, with the genotype of the −8503 G/A polymorphism in the promoter of ADIPOR1 (n=85, p=0.056 after adjustment for age, sex and percentage of body fat) in the cross-sectional study. The insert shows liver fat before (white bars) and after an intervention (black bars) with dietary counselling and an increase in physical activity in the longitudinal study. The p values (ANOVA) for liver fat at follow-up are adjusted for liver fat at baseline, sex and percentage of body fat at baseline and at follow-up
In contrast, there was no association with markers of inflammation, such as C-reactive protein and white blood cell count, insulin secretory function and the plasma adipokines TNF-α, IL-6 and adiponectin (all p>0.26, all adjusted for age, sex and percentage body fat) (Table 3). The SNPs were also not associated with intramyocellular lipid content and the plasma lipid profile (triglycerides, total, HDL cholesterol and LDL cholesterol; all p>0.40; data not shown).
In ADIPOR2, none of the alleles at the SNPs tested were associated with any anthropometrics or metabolic characteristics. Results for the alleles at the two SNPs in the promoter (−64774 T/C, −64053 G/A) and the SNP in intron 5 that was almost in complete linkage disequilibrium (D′ >0.95) with the SNPs in exon 6 and exon 7 are shown in ESM Table 1.
All results were not affected by adjustment for family membership (between-family, random effect in the multiple linear regression models). Using haplotype analyses, ten haplotypes were found in our population for ADIPOR1. The five most common haplotypes (frequency >0.02) are shown in Table 4. In ADIPOR1, an at-risk haplotype was found containing the A allele at SNP −8503 G/A and the C allele at SNP −1927 T/C. This haplotype was associated with lower insulin sensitivity estimated from the OGTT and determined during the clamp and with higher HbA1c levels compared with the so-called wild-type haplotype, which consisted only of the major alleles. This was independent of age, sex and percentage of body fat. There was no difference in the three phenotypes tested between any of the other three haplotypes compared with the wild-type haplotype. Because of the small number of subjects who had measurements of liver fat, haplotype analyses were not performed for this parameter.
Nine haplotypes were found for ADIPOR2. The three most common haplotypes (frequency >0.02) are shown in ESM Table 2. There was no significant difference in insulin sensitivity or HbA1c levels between the two haplotypes and the wild-type haplotype.
Longitudinal study
Of the 85 subjects who had measurements of liver fat at baseline, 45 had data on liver fat at follow-up and were included in the longitudinal analyses. The population consisted of 19 males and 26 females with a mean age of 44±2 years (mean±SE) and a mean BMI of 29.3±0.6 kg/m2 at baseline. Of these, 33 had NGT and 12 had IGT at baseline. The mean follow-up time was 9.8±0.5 months. Body weight decreased from 86±2 to 84±2 kg. Liver fat decreased from 5.9±0.8 to 4.5±0.7% (p=0.0002), whereas there was no significant change in insulin sensitivity (clamp, from 0.058±0.005 to 0.056±0.004% μmol kg−1 min−1 [pmol/l]−1); p=0.54) on average. There was no difference in change in body weight when subjects who were homozygous for the major alleles were compared with those who were homozygous or heterozygous for the minor alleles (−8503 G/A polymorphism in the promoter and the −1927 T/C polymorphism in intron 1; p=0.27). However, in subjects carrying the A allele (−8,503 G/A, n=27, homozygotes and heterozygotes combined) insulin sensitivity at follow-up was lower (p=0.03) and liver fat was higher (p=0.008) compared with subjects carrying the −8503 G allele (n=18, homozygotes) independent of insulin sensitivity or liver fat at baseline, sex, and percentage of body fat at baseline and at follow-up (Table 5, Fig. 2).
Discussion
In the cross-sectional study we found that the A allele at the SNP −8503 G/A in the promoter and the C allele at the novel SNP −1927 T/C in intron 1 of the ADIPOR1 gene were associated with insulin resistance. In addition, we detected a common haplotype carrying the −8503 A and the −1927 C alleles that was also associated with insulin resistance. Thus, these findings are compatible with the hypothesis that genetic variants of ADIPOR1 may play a role in glucose metabolism in humans. However, replication of these data in other populations and functional studies on these genetic variants are necessary to clarify the role of these SNPs in metabolism. Mechanisms of adiponectin action possibly include direct effects of adiponectin on lipid metabolism in the liver, resulting in lower liver fat. Ectopic fat disposition, particularly in the liver, is considered a major factor preceding insulin resistance and type 2 diabetes [25, 26]. Our findings of slightly higher liver fat in subjects carrying the −8503 A and −1927 C alleles therefore support our hypothesis that regulation of liver fat may represent a link between the polymorphisms and insulin resistance.
Of particular interest is the fact that the polymorphisms were associated with higher liver fat after an intervention to reduce adiposity and to prevent type 2 diabetes in the longitudinal study. This was independent of liver fat at baseline. In addition, the polymorphisms were associated with lower insulin sensitivity. It is of note, however, that the number of subjects included in this longitudinal study and the effects observed are rather small. Therefore, it is difficult to infer a general statement from these data with respect to the functional relevance of the polymorphisms. Nevertheless, we feel that our findings are somewhat strengthened by the fact that there was an association of the polymorphisms with these traits, not only in the cross-sectional but also in the longitudinal study. This does not represent a real replication of the data. However, with the lifestyle intervention in these individuals with a large decrease in liver fat in a short period of time, additional, more acute effects of the polymorphisms on metabolism may be detected. Of further interest is the fact that weight loss was not different between subjects who were homozygous for the major (−8 503 G and −1927 T) alleles and the heterozygotes and those homozygous for the minor alleles combined. If these findings can be replicated in other studies, one may expect that particularly subjects carrying the minor alleles may require a more intense intervention to decrease liver fat and to improve insulin sensitivity.
We want to point out that our main goal in this study was to screen for SNPs in both receptors without an a priori hypothesis. This naturally results in performing a large number of tests and we are aware that this may increase the risk of a type 1 error. One way to deal with this problem is to perform Bonferroni correction. However, it remains debatable whether one should adjust data for numbers of tests [27]. The theoretical basis for advocating adjustment for multiple comparisons is the universal null hypothesis, whereby each hypothesis tested is independent of each of the others. Because the multiple hypotheses tested here are highly correlated, we did not perform Bonferroni correction for independent comparisons. The known physiological role of ADIPOR1 and ADIPOR2 in the regulation of insulin sensitivity and ectopic fat deposition led us to believe that our findings may be relevant. Nevertheless, we readily recognise that a conservative adjustment for multiple comparisons would render all of these associations non-significant.
In contrast to ADIPOR1, polymorphisms in ADIPOR2 were not associated with liver fat, insulin sensitivity or any parameter of lipid metabolism and inflammation. Performing haplotype analyses, we could also not detect an association of a haplotype with, for example, insulin sensitivity, the phenotype we were interested in most. Clearly, we can only speculate that variants of this gene do not play a major role in our population. The impact of ADIPOR2 on glucose and lipid metabolism, however, may still be important.
We also found that the A allele at the SNP −8503 G/A and the C allele at the SNP −1927 T/C of ADIPOR1 were associated with higher plasma HbA1c levels. One may expect that the genotypes for the polymorphisms may also be differently distributed in subjects with NGT and IGT. However, genotype distribution was not different between the two groups (p=0.60, χ 2 test). An explanation may be that our group of subjects with IGT was rather small and a difference may not be easily detected using this dichotomous variable. Because the associations of the polymorphisms with the phenotypes were similar in the two groups, subjects with NGT and IGT were pooled for these analyses.
Having found that the SNP in the promoter and one SNP in intron 1 of ADIPOR1 were associated with insulin resistance, increased liver fat and higher HbA1c levels, the question arose whether these SNPs may be functional or whether they are in linkage disequilibrium with a yet unidentified polymorphism. We screened the public TRANSFAC database [28] for transcription factors that bind to these sites. We identified a zinc finger protein (IKAROS; IK1) that binds to the wild type but not to the mutant sequence of the promoter at position −8503. This protein, which was identified in the late 1990s, plays a role in lymphocyte differentiation [29]. Whether it is also relevant in glucose and lipid metabolism is not known. For the −1927 T/C SNP in intron 1, which is in complete linkage disequilibrium with the promoter polymorphism, there are no transcription factors known that may bind to this site. Because we identified several putative binding sites for transcription factors in the TRANSFAC-database, such as C/EBP, CREB and HNF-4, this intron may function as an enhancer of transcription. Thus, another (not yet identified) SNP in intron 1 in linkage disequilibrium with the −1927 T/C SNP may be functional.
Most recently, the group of Elbein found that SNPs in ADIPOR1 were not associated with type 2 diabetes [24]. Nevertheless, two alleles in the 3′ untranslated region were expressed unequally and mRNA of ADIPOR1 were lower among transformed lymphocytes from diabetic African-American individuals compared with healthy controls. This was interpreted as a possible role of ADIPOR1 in the metabolic syndrome. Among the SNPs found in our population, the −8503 G/A SNP in the promoter and the 5843 T/C SNP in intron 4 were also detected in that study. Although the A allele at the SNP −8503 G/A was associated with BMI in interaction with pedigree membership, no association of the SNP with insulin sensitivity was found in individuals of Northern European descent. The different findings possibly result from the smaller number of subjects (n=126) included in that analysis.
The group of Shuldiner also screened both genes [30]. They found five SNPs in ADIPOR1. Among them, two SNPs (−8503 G/A in the promoter and 5843 T/C in intron 4) were also found in our population. The other three SNPs were located in regions that were not screened in our population. While the SNP in the promoter was not associated with type 2 diabetes (p=0.17), the SNP in intron 4 showed a significant association. Sixteen SNPs were found in ADIPOR2. Among them, SNPs in the promoter, in intron 2 and in the 3′ untranslated region were associated with type 2 diabetes. We cannot explain the different findings compared with our study. Different statistical approaches and different ethnic homogeneities of the populations, resulting in different allelic frequencies, may play a role here.
In conclusion, we found that variants of ADIPOR1 were associated with insulin resistance and high liver fat in humans. This supports the hypothesis that adiponectin, as shown for animals, may also regulate liver fat and insulin sensitivity in humans. Whether these polymorphisms also affect type 2 diabetes needs to be investigated in epidemiological studies.
Abbreviations
- ADIPOR:
-
adiponectin receptor
- ESM:
-
electronic supplementary material
- EMCL:
-
extramyocellular lipid content
- IMCL:
-
intramyocellular lipid content
- SNP:
-
single-nucleotide polymorphism
References
Maeda N, Shimomura I, Kishida K et al (2002) Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med 8:731–737
Kubota N, Terauchi Y, Yamauchi T et al (2002) Disruption of adiponectin causes insulin resistance and neointimal formation. J Biol Chem 277:25863–25866
Yamauchi T, Kamon J, Waki H et al (2001) The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med 7:941–946
Berg AH, Combs TP, Du X, Brownlee M, Scherer PE (2001) The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med 7:947–953
Fruebis J, Tsao TS, Javorschi S et al (2001) Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci USA 98:20005–20010
Combs TP, Berg AH, Obici S, Scherer PE, Rossetti L (2001) Endogenous glucose production is inhibited by the adipose-derived protein Acrp30. J Clin Invest 108:1875–1881
Chandran M, Phillips SA, Ciaraldi T, Henry RR (2003) Adiponectin: more than just another fat cell hormone? Diabetes Care 26:2442–2450
Goldfine AB, Kahn CR (2003) Adiponectin: linking the fat cell to insulin sensitivity. Lancet 362:1431–1432
Stefan N, Vozarova B, Funahashi T et al (2002) Plasma adiponectin concentration is associated with skeletal muscle insulin receptor tyrosine phosphorylation, and low plasma concentration precedes a decrease in whole-body insulin sensitivity in humans. Diabetes 51:1884–1888
Yamamoto Y, Hirose H, Saito I, Nishikai K, Saruta T (2004) Adiponectin, an adipocyte-derived protein, predicts future insulin resistance: two-year follow-up study in Japanese population. J Clin Endocrinol Metab 89:87–90
Lindsay RS, Funahashi T, Hanson RL et al (2002) Adiponectin and development of type 2 diabetes in the Pima Indian population. Lancet 360:57–58
Spranger J, Kroke A, Möhlig M et al (2003) Adiponectin and protection against type 2 diabetes mellitus. Lancet 36:226–228
Yamauchi T, Kamon J, Ito Y et al (2003) Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 423:762–769
Civitarese AE, Jenkinson CP, Richardson D et al (2004) Adiponectin receptors gene expression and insulin sensitivity in non-diabetic Mexican Americans with or without a family history of type 2 diabetes. Diabetologia 47:816–820
Tsuchida A, Yamauchi T, Ito Y et al (2004) Insulin/Foxo1 pathway regulates expression levels of adiponectin receptors and adiponectin sensitivity. J Biol Chem 279:30817–30822
Weiss R, Dufour S, Taksali SE et al (2003) Prediabetes in obese youth: a syndrome of impaired glucose tolerance, severe insulin resistance, and altered myocellular and abdominal fat partitioning. Lancet 362:951–957
Tiikkainen M, Tamminen M, Hakkinen AM et al (2002) Liver-fat accumulation and insulin resistance in obese women with previous gestational diabetes. Obes Res 10:59–87
Kelley DE, McKolanis TM, Hegazi RA, Kuller LH, Kalhan SC (2003) Fatty liver in type 2 diabetes mellitus: relation to regional adiposity, fatty acids, and insulin resistance. Am J Physiol Endocrinol Metab 285:906–916
Expert Committee on the Diagnosis and Classification of Diabetes Mellitus (1997) Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 20:1183–1197
Matsuda A, DeFronzo R (1999) Insulin sensitivity indices obtained from oral glucose tolerance testing. Diabetes Care 22:1462–1470
Stumvoll M, Mitrakou A, Pimenta W et al (2000) Use of the oral glucose tolerance test to assess insulin release and insulin sensitivity. Diabetes Care 23:295–301
Thamer C, Machann J, Bachmann O et al (2003) Intramyocellular lipids: anthropometric determinants and relationships with maximal aerobic capacity and insulin sensitivity. J Clin Endocrinol Metab 88:1785–1791
Tregouet DA, Escolano S, Tiret L, Mallet A, Golmard JL (2004) A new algorithm for haplotype-based association analysis: the Stochastic-EM algorithm. Ann Hum Genet 8:165–177
Wang H, Zhang H, Jia Y et al (2004) Adiponectin receptor 1 gene (ADIPOR1) as a candidate for type 2 diabetes and insulin resistance. Diabetes 53:2132–2136
Danforth E Jr (2000) Failure of adipocyte differentiation causes type II diabetes mellitus? Nat Genet 26:13
Angulo P (2002) Nonalcoholic fatty liver disease. N Engl J Med 346:1221–1231
Rothman KL (1990) No adjustments are needed for multiple comparisons. Epidemiology 1:43–46
Matys V, Fricke E, Geffers R et al (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31:374–378
Dumortier A, Kirstetter P, Kastner P, Chan S (2003) Ikaros regulates neutrophil differentiation. Blood 101:2219–2226
Damcott C, Ott SH, Pollin GI et al (2004) Genetic variation in adiponectin receptor 1 (ADIPOR1) and adiponectin receptor 2 (ADIPOR2) is associated with type 2 diabetes (T2DM) in the Old Order Amish. Diabetes 54:2245–2250
Acknowledgements
We thank all the research volunteers for their participation. We gratefully acknowledge the superb technical assistance of E. Maerker, A. Teigeler, H. Luz, M. Weisser, A. Guirguis and C. Peterfi. The authors would like to thank all participants for their cooperation.
The study was supported in part by a grant from the German Research Foundation (Deutsche Forschungsgemeinschaft, KFO 114/1) and the European Community’s FP6 EUGENE 2 (LSHM-CT-2004-512013).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Stefan, N., Machicao, F., Staiger, H. et al. Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia 48, 2282–2291 (2005). https://doi.org/10.1007/s00125-005-1948-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-005-1948-3