Abstract
Background
The genus Limonium Miller comprises annual and perennial halophytes that can produce sexual and/or asexual seeds (apomixis). Genetic and epigenetic (DNA methylation) variation patterns were investigated in populations of three phenotypically similar putative sexual diploid species (L. nydeggeri, L. ovalifolium, L. lanceolatum), one sexual tetraploid species (L. vulgare) and two apomict tetraploid species thought to be related (L. dodartii, L. multiflorum). The extent of morphological differentiation between these species was assessed using ten diagnostic morphometric characters.
Results
A discriminant analysis using the morphometric variables reliably assigns individuals into their respective species groups. We found that only modest genetic and epigenetic differentiation was revealed between species by Methylation Sensitive Amplification Polymorphism (MSAP). However, whilst there was little separation possible between ploidy levels on the basis of genetic profiles, there was clear and pronounced interploidy discrimination on the basis of epigenetic profiles. Here we investigate the relative contribution of genetic and epigenetic factors in explaining the complex phenotypic variability seen in problematic taxonomic groups such as Limonium that operate both apomixis and sexual modes of reproduction.
Conclusions
Our results suggest that epigenetic variation might be one of the drivers of the phenotypic divergence between diploid and tetraploid taxa and discuss that intergenome silencing offers a plausible mechanistic explanation for the observed phenotypic divergence between these microspecies. These results also suggest that epigenetic profiling offer an additional tool to infer ploidy level in stored specimens and that stable epigenetic change may play an important role in apomict evolution and species recognition.
Similar content being viewed by others
Background
DNA sequence divergence clearly plays a leading role in shaping the phenotypic variation observed between most taxa e.g. [1, 2] but does not explain all forms of adaptive phenotypic differentiation [3–5]. Epigenetic modifications of DNA and histones, the core components of chromatin are able to influence the expression of the underlying genes and so phenotype [6]. Of these epigenetic mechanisms, the best studied and the one to show more examples of transgenerational stability is cytosine DNA methylation [6, 7]. Evidence has also been found suggesting that heritable phenotypic variation observed in natural populations can be due to stable epigenetic variation, even in the absence of genetic variation and could play a role in plant adaptation and evolution [5, 8–10]. The scope for epigenetic mechanisms driving morphological differentiation is perhaps best illustrated when genetic variation is lacking. Understanding the relative importance of genetic and epigenetic sources of phenotypic variation where both systems are operating is therefore attracting increasing interest. For example, Laguncularia racemosa is a mangrove plant species that shows low genetic variability but in populations from distinct, nearby habitats, cytosine methylation variation among individuals correlates more closely with environmental variation than does genetic variation [11]. In the perennial Viola cazorlensis, cytosine methylation patterns were found to be partitioned and positively correlated with adaptive genetic variation [12]. Also, in populations of individuals with reduced or negligible genetic variation such as those of triploid asexual dandelion lineages (apomixis; diplospory), changes in genomic methylation patterns are found between individuals [13].
The genus Limonium Miller (sea-lavenders; Plumbaginaceae) has long been recognized to have a history of recurrent hybridization and polyploidization, and comprises 150 [14] to 350 taxa recognized across coastal, steppe and desert regions (e.g. [15, 16]). This wide range is due to the description of new taxa, mainly microspecies from geographically restricted areas. In this genus, a sporophytic self-incompatibility system is linked with pollen-stigma dimorphisms, A-pollen type grains germinate on papillose stigmas and B-pollen type germinate in cob-like stigmas, while the complementary combinations produce no successful fertilization [17, 18]. Most sexual species of Limonium usually have a dimorphic self-incompatibility system (both pollen and stigmas are dimorphic) while agamospermous species are generally monomorphic and have monomorphic populations [14, 19]. Determination of these characters in individuals from natural and/or experimental populations has since long been used as an indirect method for estimation of each species reproduction mode [14, 17, 18]. The high number of polyploid taxa has been explained to be a consequence of this self-incompatibility system and the ability of polyploid hybrids to produce seeds asexually via apomixis (agamospermy; asexual seed formation) [14, 17–20]. Ixeris-type embryo sacs with non-haploid eggs are found in triploid (2n = 3x = 27) Statice oleaefolia var. confusa[20]. In triploid and tetraploid agamospermous species of the L. binervosum (G. E. Sm.) Salmon group diplospory followed by parthenogenesis is reported [21, 22]. Molecular phylogenetic studies have tried to resolve the taxonomic complexity within this genus in a global perspective using nuclear DNA sequence information [23] and plastid DNA [24–26].
In Continental Portugal about 15 Limonium species have been recognized with ecological importance for plant communities of the Atlantic and Mediterranean coastlines [15, 27]. Among these, the L. ovalifolium complex consists a group of three sexual diploids (2n = 2x =16): L. ovalifolium (Poir.) O. Kuntze, L. nydeggeri Erben and L. lanceolatum (Hoffmanns & Link) Franco [28]. The first species has a broader distribution including several Sites of Community Importance (SCI) for the Mediterranean biogeographical regions [29] in the West (Estremadura), Southwest Alentejo and Algarve coastlines [15, 30]. Conversely, the Lusitania endemic L. nydeggeri and L. lanceolatum have more restricted distributions; the former is restricted to West and Southwest Atlantic sea-cliffs whereas the latter is found in the Southwest and South coastlines [28, 30]. Limonium tetraploid taxa include among others, the Lusitania endemic apomict, L. multiflorum Erben [14, 15] which exhibits both tetraploid and aneuploid tetraploid cytotypes (2n = 4x = 35 - [14]; 2n = 4x = 32, 34, 35, 36 – [31]) and the aneuploid tetraploid apomict L. dodartii (Girad) O. Kuntze (2n = 4x = 35) which most frequently grow on maritime cliffs in the province of Estremadura. A third tetraploid, L. vulgare (2n = 4x = 36), a sexual species [19, 32, 33], grows in salt marshes [15, 30].
Dominant genetic markers, such as those generated by Amplified Fragment Length Polymorphism (AFLP), are valuable for assessing genetic diversity within and between populations [34] and for inferring taxon differentiation [35], especially in species for which codominant markers are unavailable. Some recent publications have added data on natural epigenetic variation in animal and plant species by sampling cytosine methylation using Methylation-Sensitive Amplification Polymorphism (MSAP) technology [36–38], a modification to the original AFLP protocol that compares product profiles generated by methylation-sensitive/insensitive isoschizomers. Central to the technique is the differential behavior of the two isoschizomer restriction enzymes (HpaII and MspI) in the presence of cytosine methylation in the CCGG context. HpaII is inactive if one or both cytosines are methylated at both DNA strands, but cleaves when one or both cytosines are methylated in only one strand. MspI, by contrast, cleaves C5mCGG but not 5mCCGG. Comparison of the profiles generated by each enzyme from each individual allows the assessment of the methylation state of the restriction sites and so provides a relative comparison of genetic and epigenetic variability. Several reports suggest that only methylation marks in the CG context can be transmitted between generations and so have potential for stable adaptive significance [39]. Furthermore, recent data shows that although non-CG methylation can be inherited, only inherited CG methylation is inversely correlated with gene expression [40]. Since only HpaII is affected by methylation of this kind, for simplicity, in this study we refer to profiles from this enzyme as epigenetic (meaning potentially transgenerationally stable and epigenetic) whereas MspI is insensitive in this sense and so can only detect transgenerationally relevant genetic variation. The focus of this study was to therefore to use MSAP analysis as the primary tool in comparing the extent to which genetic and epigenetic diversity in natural populations of diploid and tetraploid Limonium species correlate with species identity and ploidy.
Results
Morphological differentiation between diploid and tetraploid species
Herbarium specimens of the diploid species L. ovalifolium, L. nydeggeri and L. lanceolatum and of the tetraploid species L. dodartii, L. multiflorum and L. vulgare obtained from individuals sampled in natural populations were used for morphometric measurements. Ten diagnostic characters were selected based on an exhaustive review of Limonium species in Southwest Europe by Erben [14, 15], and on previous biometric studies in the Limonium genus [33, 41, 42].
Only one of the ten morphological variables measured from representatives of the six species fitted a normal distribution, Maximum inner bract length (MIBL), whilst the other nine failed to do so, even after a logarithmic transformation. The remaining analyses were therefore performed using the original (untransformed) values (Additional file 1). Canonical discriminant analysis (CDA) of the morphological variables accounted for most of the variation (82.2% in the first two dimensions, comprising 61.9 and 20.3%, respectively) and correctly assigned individuals to species in 92.7% of cases (n = 110; Figure 1; Table 1). However, a small number of intermediate or ambivalent specimens were encountered (Wilks’ lambda = 0.002, χ 2 = 650.259 > χ20.05;50 = 67.505, P < 0.001). The first axis distinguished diploid from tetraploid species through the following characters: Maximum outer bract length (MOBL), Maximum calyx length (MCL), Maximum middle bract length (MMBL) and MIBL (Table 2; Figure 2). The second axis separated L. nydeggeri from the other diploid species and L. multiflorum from the other tetraploid species by the following characters: Maximum middle bract width (MMBW), Maximum outer bract width (MOBW) and Maximum inner bract width (MIBW). Thus, these features were largely responsible for separation of species sharing the same ploidy level.
Discriminant function analyses of morphometric data with predefined diploid and tetraploid Limonium species. Individuals from each diploid (L. lanceolatum, L. nydeggeri, L. ovalifolium) or tetraploid (L. dodartii, L. multiflorum and L. vulgare) species are represented by colour symbols. Each species centroid is represented by filled squares. Percentages of total variance explained by the functions are given in parentheses.
Box plots of significant characters that discriminate diploid from tetraploid Limonium species. The box from diploids L. nydeggeri, L. ovalifolium, L. lanceolatum, and tetraploids L. dodartii, L. multiflorum, L. vulgare show the twenty-fifth and seventy-fifth percentile ranges and the median; circles and asterisks are outliers (cases with values between 1.5 and 3 box lengths from the upper or lower edge of the box). MOBL - Maximum outer bract length; MCL - Maximum calyx length; MMBL - Maximum middle bract length; and MIBL - Maximum inner bract length.
MSAP profiles analysis
Genetic-epigenetic analyses were performed on 125 individuals selected from natural populations within designated SCI(s) (Figure 3; Table 3). The two MSAP primer combinations applied to all samples yielded 835 scorable fragments comprising 792 from MspI and 778 from HpaII, 92.78% and 95.36% respectively, were polymorphic (i.e. not present in all the analysed samples/replicates when restricted with one of the isoschizomers). Overall reproducibility between biological replicates was 83% and 85% for primer combinations E1/H1 and E1/H3 respectively. The methylation insensitive (genetic variation) profiles showed only very slightly higher concordance among replicates (83.6% and 85.7%) than did the methylation-sensitive (epigenetic variation) profiles (82.4% and 84.3%). Technical reproducibility of the MSAP technique revealed between 92-95% band concordances (data not shown), indicating that the higher variability between independent DNA extractions probably arises from variation between tissues and cell mixtures used in the DNA extraction.
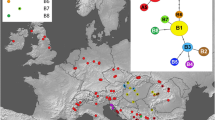
Diploid and tetraploid Limonium populations sampled in continental Portugal in Sites of Community Interest. Diploid species (L. lanceolatum, L. nydeggeri and L. ovalifolium) are represented by full stars and tetraploid species (L. dodartii, L. multiflorum and L. vulgare) are represented by full circles. Specimens were sampled in Estremadura (PTCON0056: Baleal, Papoa, Nossa Sra dos Remédios, Vale dos Frades, Foz do Lizandro; PTCON0008: Cabo Raso), Alentejo (PTCON0012: Vila Nova de Milfontes, Pontal da Carrapateira), and in Algarve (PTCON0012: Cabo de Sagres; Praia da Luz) provinces.
Profiles from the tetraploid species (i.e. L. multiflorum, L. dodartii, L. vulgare) included a higher number of MspI fragments (genetic profiles) per individual for both primer combinations (Table 4). Conversely, these species contained a lower number of HpaII-generated (methylation-sensitive epigenetic profiles) and fewer fragments per individual than the three diploid species (i.e. L. nydeggeri, L. ovalifolium, L. lanceolatum), implying a higher level of genome-wide methylation among tetraploids.
Genetic/epigenetic divergence of diploid and tetraploid species
Principal Coordinate Euclidean Analysis (PCoA) was used to provide an overview of the genetic/epigenetic variability and structure of the studied taxa. Overall, epigenetic profiles (HpaII) provided imperfect but slightly better separation of the taxa than did the genetic profiles (MspI) (Figure 4A; Additional file 2). Genetic data (MspI) alone also largely failed to discriminate within the diploid species and the tetraploid species (Figure 4B; Additional file 2). More remarkably, PCoA plots of the epigenetic HpaII profiles revealed clear separation between the diploid and tetraploid taxa (Figure 4C; Additional file 2).
Principal Coordinate Analysis (PCoA) representing genetic and epigenetic variability in diploid and tetraploid Limonium species. PCoA was based on presence/absence scores of 488 polymorphic loci obtained from MSAP profiles using isoschizomers MspI (methylation insensitive - red symbols in A and B) or HpaII (methylation sensitive - blue symbols in A and C) as frequent cutters and amplified with primers (E1/H3). The first two coordinates were extracted and plotted against each other. Percentage of the variability shown by each coordinate is indicated between parentheses. Diploid species are represented by solid symbols (L. lanceolatum, triangles; L. nydeggeri, rhomboids; L. ovalifolium, rectangles) and tetraploid species are represented by empty symbols (L. dodartii, triangles; L. multiflorum, rectangles; L. vulgare, circles).
On average, genetic and epigenetic distances between the three diploid species were significantly lower than comparable distances between the tetraploid species (T-test, Genetic distance p < 0.023; epigenetic distance p < 0.0006 (two-tailed test); Additional file 3). Furthermore, calculated epigenetic distances between individuals from different diploid species and between tetraploid species were higher than genetic distances between the same pairings (T-test, p < 0.04 (between diploid species); p < 0.15 (between tetraploid species) (two-tailed test); Additional file 3).
Analysis of genetic/epigenetic variability using Analysis of Molecular Variance (AMOVA) showed that 24% of the total observed variability can be attributed to differences between epigenetic (HpaII) and genetic (MspI) sources of variability (Figure 4A; Additional file 2). Independent analysis of results generated by each enzyme type revealed that differences between diploid and tetraploid samples accounted for 6-8% of the genetic variability and for 3-6% of the epigenetic variability. This compared with a much higher level of within-species variation, which comprised 79-83% and 74-78% of the genetic and epigenetic variability respectively (Figure 4B-C; Additional file 2). Surprisingly, while genetic differences between species accounted for 9-15%, epigenetic differences accounted for 19-20%, suggesting that characterisation of these very closely related species is best served by considering both genetic and epigenetic information rather than genetic information only (Additional file 3).
Tetraploid species compensate lower genetic variability with higher epigenetic variability
Closer consideration of the genetic and epigenetic variability revealed that all species had more epigenetic than genetic variability. And, the diploid species were more variable (both at genetic and epigenetic level) than were the tetraploid ones (Table 5). However, the difference between genetic and epigenetic variability was higher among the tetraploid species despite containing lower levels of variability overall (Table 5).
PCoA of the genetic profiles revealed no or only weak co-clustering of individuals according to population origin, with the best example being seen among L. nydeggeri samples (Figure 5 A-B). No co-clustering was evident from the epigenetic profiles (data not shown). A lack of structuring according to population origin was further supported by AMOVA, with the main component of both genetic (90-97%) and epigenetic variance (86-95%) residing within populations.
Principal Coordinate Analysis (PCoA) representing genetic variability in Limonium nydeggeri populations. PCoA was based on presence/absence scores of 488 (primer I, E1/H1) (A) and 347 (primer II, E1/H3) (B) polymorphic loci obtained from MSAP profiles using isoschizomers MspI (methylation insensitive) or HpaII (methylation sensitive) as frequent cutters. The first two coordinates were extracted and plotted against each other.
We next sought structuring across a geographic scale. Mantel test analysis revealed a correlation between genetic distances and geographic separation among conspecific populations for L. nydeggeri (R2 = 0.784, P < 0.03). This correlation was significant for both primer pairs (H1/E1, R2 = 0.858, P < 0.02; H1/E3, R2 = 0.616, P < 0.04) (Figure 6 A-B). Both primers generated a scatterplot showing a positive and monotonic relationship over all geographic distances of separation. In contrast, L. multiflorum populations showed no detectable relationship between genetic distance and geographic distance and large variance in estimates of divergence. None of the studied species showed a significant correlation between epigenetic and geographic distances, with extensive scatter between the plotted samples (data not shown).
Correlation between pairwise genetic differentiation (GD, PhiPT) and geographical distance (GGD, in Km) (A-B) and between pairwise genetic and epigenetic differentiation (GD and EpiGD, respectively) (C) between Limonium nydeggeri populations. Mantle tests were based on MSAP data obtained using HpaII and primer combinations primer I (E1/H1) (A) and primer II (E1/H3) (B) and HpaII and MspI and both primers (C). Shown equations are the linear functions and R2 values for each Mantel test. Analysis using 1000 permutation tests showed a significant correlations (A: P < 0.04; B: P < 0.02; C: P < 0.03).
Finally, Mantel test analysis revealed a strong significant positive correlation between genetic and epigenetic distances among L. nydeggeri populations (H1/E1, R2 = 0.800, P < 0.008; H1/E3, R2 = 0.200, P < 0.07 and R2 = 0.600, P < 0.005, using the information provided by both primer combinations) (Figure 6 C). Conversely, there was only a weak negative correlation between genetic and epigenetic distances among L. multiflorum populations (data not shown).
Discussion
The effects of hybridization, polyploidy and apomixis have all combined to shape radiation currently seen in Limonium species [26, 31, 43]. This evolutionary model has been used to explain multiple series of complex aggregates of sexual diploid species and asexual polyploid hybrids which are perpetuated through gametophytic apomixis [44–48]. In other plant groups, hybridization and polyploidy have combined to generate genetic and phenotypic complexity in the form of classical polyploidy pillar complexes. In these situations species delimitations typically become blurred at the higher ploidy levels and interploidy discrimination can also become difficult for some taxa. The many examples of this type of species complex include the Festuca ovina aggregate [49], Dactylis glomerata[50] and Knautia arvensis species groups [51]. The additional and intermittent appearance of facultative or obligatory apomixis, as seen in triploid and tetraploid Limonium species [20, 21], adds another layer of complexity for species delimitation and diagnosis. In these instances the phenotypic range of taxa can be highly variable, as can their morphological distinctiveness and stability (e.g. [52]). The cumulative effect of these processes is typically manifest in the recognition of a large number of microspecies, with the diagnosis and genetic characterisation of many of the resultant taxa often being highly demanding (e.g. Limonium, reviewed in [26]). The publication of a comprehensive revision of Limonium species in Southwest Europe by Erben [14] in which 59 species (13 new to science) were fully described allowed for several taxometric studies of geographically related species in this genus to be conducted. For example, various authors used a taxometric approach to study several sexual and agamospermous species of the L. binervosum (G. E. Sm.) C. E. Salmon complex from Western Europe [53], and also to examine the closely related L. vulgare and L. humile species in the British Isles [33]. Morphometric studies have also been used at population level. For instance, some authors studied the taxometric relationships between triploid or aneuploid tetraploid L. binervosum agamospermous colonies in the British Isles [42, 53]. A similar strategy is applied to describe morphological differentiation patterns between individuals of a L. dufourii population from Eastern Spain [54].
In the current study, CDA was applied using ten morphometric traits collected from representative individuals of three diploid and three tetraploid Limonium species from Portugal. As previous studies had suggested [14, 33, 41, 42], the collective use of these characters in CDA was sufficient to provide clear morphological differentiation between species at each ploidy level. This differentiation is based primarily on the use of seven morphometric variables, viz: MOBL, MCL, MMBL and MIBL for the first axis and MMBW, MOBW and MIBW on the second. Overall, these analyses not only provided clear separation of all diploid species but also indicated that L. lanceolatum and L. ovalifolium share a closer phenotypic affinity. Separation of the tetraploid species was also possible but with intermediate individuals blurring the boundaries between L. vulgare and L. dodartii, and between the latter and L. multiflorum. The rather surprising finding, however, lay in the clear phenotypic separation of tetraploid species from the diploids on the basis of the bract and calyx characteristics MCL, MOBL, MMBL and MIBL rather than plant size features more usually associated with ploidy level changes (e.g. see [55]). The relative importance of genetic and epigenetic processes in shaping the observed phenotype structuring among species and ploidy levels in Limonium has thus far remained elusive.
Previous works on the analysis of genetic variation and population genetic structure in other Limonium species have deployed a wide range of molecular marker systems to infer the importance of genetic structuring in defining interspecies delimitation. There has, nevertheless, been considerable evidence that modest but significant levels of genetic variation does occur within species in the genus. For example, studies on the presumed agamospermous triploid L. dufourii have invariably revealed low but substantial inter-individual genetic variation in habitats with significant fragmentation and low population sizes [54, 56–58]. Similarly, in diploid L. dendroides from Canary Islands, despite radical habitat fragmentation and small population size, some subpopulations of have enough genetic variation to compensate for the influence of drift [59]. However, in plant populations with low genetic variability, epigenetic variation can also act as an important source of potentially adaptive phenotypic variability [11–13]. Nevertheless, the extent and importance of epigenetic variation in natural populations of sexual and agamospermous Limonium species is still largely unexplored [9, 10, 60].
Data generated in the current study unsurprisingly reveals that tetraploids have a higher level of methylation than diploids. In part this may be accounted for the increased genome size, with scope for intergenome heterozysity for methylation marks adding to that expected between homologous loci. There are also additional indirect processes that can contribute to an expected increased incidence in methylation across polyploid species. In newly formed polyploids, genomic duplications from transposons and duplication of regulatory genes, can result in high levels of cytosine methylation on the new portions of the genome [61–63]. Several methylation-based processes have been implicated in the competitive silencing of duplicated gene regions (e.g. [64]), leading to increased methylation relative to the diploid progenitor(s). The same processes also lead to the expectation of differential methylation patterning between the diploid progenitor and the polyploid offspring, partly because of imperfect duplication of genes in the neopolyploid, as was reported for the Waxy gene in Spartina[64], but also because of various systems of asymmetric homeolog silencing (e.g. [64]). Given the causal links between methylation and gene regulation [5], there is scope for such changes to influence the phenotype of the polyploids relative to their diploid relatives. Principal coordinate and genetic distance analyses performed in the current study yielded greater separation between diploid and tetraploid taxa when using epigenetic information than when using only genetic information, suggesting that ploidy levels are better separated using epigenetic information than genetic information alone. Intriguingly, this pattern of variation was mirrored by the more pronounced morphological separation between the diploid and tetraploid taxa than that seen between species at either ploidy level. Should this observation apply more broadly to other groups, it implies that epigenetic profiling may provide a useful additional tool to infer ploidy level of preserved specimens.
We found similar trends when making comparisons at the species level. Moreover, the diploid species within L. ovalifolium complex have imperfect but reasonable morphological differentiation but genetic co-variation with species identity was relatively modest. Interspecies separation was more strongly enhanced when analysis was focused on the epigenetic variation encompassed in the HpaII profiles, implying that epigenetic patterning (and associated gene silencing) may play a significant role in species separation of the group and so may have some utility for species diagnosis. In both cases, further research would be required to convert these anonymous profiles into Sequence Tagged Site epigenetic markers for robust diagnostic purposes.
Analysis of genetic/epigenetic variability using AMOVA revealed that tetraploid species present lower levels of genetic variability than diploid species. This observation could be most plausibly explained through consideration of their reproductive biology. Several previous studies of the diploids L. ovalifolium, L. nydeggeri, and the tetraploids L. dodartii, L. multiflorum and L. vulgare have provided some insights into their primary reproductive strategies [14, 31, 65]. These and other works were based on the determination of flower dimorphisms linked to a sporophytic self-incompatibility system [14, 17, 18, 28, 33, 66, 67]. In these studies diploids L. nydeggeri and L. ovalifolium were deemed probable sexual species based on their reproductive characteristics. The same applies to the tetraploid L. vulgare[14, 65]. Conversely, tetraploids L. dodartii and L. multiflorum both belonging to the Limonium binervosum group, were considered as agamospermous [31, 42, 53]. Hence, it seems most likely that the lower level of genetic diversity in the putative agamospermous tetraploids could be best explained by their apomictic reproduction mode. In other polyploid apomictic species, such as in Ranunculus sp., genetically uniform populations have been similarly observed as a consequence of this mode of reproduction [45]. This finding contrasts with the relatively high level of epigenetic variability among the tetraploids, leading us to speculate that in apomictic polyploid Limonium species, the lack of genetic variability caused by the loss of meiotic segregation could be partially compensated by enhanced epigenetic variation. Several authors have suggested similar heritable phenotypic variation due to stable epigenetic variation in the absence of genetic variation (e.g., Viola cazorlensis; Laguncularia racemosa, and in triploid asexual dandelion lineages (reviewed in [8–10])). Viewed in this context, the results of the present study add Limonium to that list.
The present work failed to support any relationship between genetic or epigenetic distance with geographic distance between populations of each species, except for a positive correlation between genetic and geographic distances among L. nydeggeri, consistent with regional equilibrium between gene flow and drift [68]. The absence of co-correlation is not unexpected for the apomictic polyploids, and can be explained by restricted gene flow between populations, founder events produced by a limited number of individuals, absence of recombination and spread of single asexual clones within populations [45]. One plausible explanation of these results considered collectively is to propose that genetic information flows between populations but that epigenetic information is mainly induced locally by the environment. Alternatively, it might be that selection pressure on epiloci is higher than on genetic loci, or simply that epiloci are plastic, in the sense that they appear and disappear depending on the environmental cues. Conversely, Mantel test analysis of the correlation between genetic and epigenetic distances between L. nydeggeri populations, which presents a regional equilibrium at genetic level, show a strong significant positive correlation between genetic and epigenetic distances. While, genetic and epigenetic distances between L. multiflorum populations, which does not present genetic regional equilibrium, showed a weak negative correlation. Again, this might suggest that on the absence of a strong gene flow between populations, environmental conditions exert a higher pressure on the fixation of epigenetic loci that cannot be masked by genetic variability introduced by sexual reproduction.
Conclusions
Higher correlation was found between morphometric and epigenetic differentiation than between morphometric and genetic differentiation. We therefore, suggest that epigenetic variation might be a driver of the observed phenotypic divergence between the studied taxa through intergenome silencing. We argue that the present work helps to demonstrate the importance of considering phenotypic, genetic and epigenetic variables when seeking to explain the dynamics of complex plant groups that feature hybridization, polyploidy and variable modes of reproductive biology.
Methods
Study species
Natural populations of the three diploid species from the L. ovalifolium complex (L. ovalifolium, L. nydeggeri, L. lanceolatum) [28, 30] and three tetraploids species (L. dodartii, L. multiflorum, L. vulgare) [14, 15] were surveyed in the three Portuguese provinces of Estremadura (West), Alentejo (South-West) and Algarve (South). With the exception of L. lanceolatum and L. vulgare (which grow in salt marshes), all populations vegetated limestone sea-cliffs, in crevices within exposed rocks or on shallow soil above the rock strata and on scree slopes where competition with other species is very low. The locations of all populations were recorded using Global Positioning System. Google Earth 6.0.2 was used for georeferencing and to estimate geographic distances between populations. Geographic mapping of the populations was performed using ArcGIS Desktop 10 (ESRI).
Three leaves per individual were sampled from approximately ten individuals for each population, with a total of 125 plants included in this study. Most individuals were selected from populations within designated SCI(s), namely in SCI Peniche/Sta Cruz (PTCON0056), SCI Sintra/Cascais (PTCON0008) and SCI Costa Sudoeste (PTCON0012) [29]. In all, five L. nydeggeri populations were surveyed from Ilha do Baleal (Estremadura: Peniche; SCI PTCON0056), Nossa Srª dos Remédios (Estremadura: Peniche SCI PTCON0056), Cabo Raso (Estremadura: Cascais; SCI PTCON0008), Pontal da Carrapateira (Algarve: Aljezur; SCI PTCON0012), Cabo de São Vicente (Algarve: Vila do Bispo; SCI PTCON0012); two L. ovalifolium populations from Cabo de Sagres (Algarve: Vila do Bispo; SCI PTCON0012), and Praia da Luz (Algarve: Lagos); one L. lanceolatum population from Vila Nova de Mil Fontes (Alentejo: Odemira; SCI PTCON0012); one L. dodartii population from Cabo Sardão (Alentejo: Odemira; SCI PTCON0012); four Limonium multiflorum populations from Península da Papoa (Estremadura: Peniche; SCI PTCON0056), Vale dos Frades (Estremadura: Lourinhã; SCI PT PTCON0056), Foz do Lizandro (Estremadura: Mafra; SCI PTCON0056) and Cabo Raso (Estremadura: Cascais; SCI PTCON0008); and one L. vulgare population from Vila Nova de Mil Fontes (Alentejo: Odemira; SCI PTCON0012) (see Figure 3; Table 3). Three leaves from plants at the same phenological stage were sampled from all sites during 2010 and kept on silica gel. In this way, variation in DNA methylation profiles attributable to developmental or storage conditions differences was minimized.
Morphometric analysis
Morphometric analyses were performed in approximately twenty herbarium specimens from each species deposited in the herbaria João de Carvalho e Vasconcellos (LISI; ISA), Portugal. These specimens were previously collected in the same populations selected for MSAP analysis and identified on the basis of species descriptions, diagnostic keys, and locations already described [14], and by comparison with other herbarium specimens present in Portuguese herbaria. The following diagnostic characters were measured: maximum spike length (MSL), maximum number of spikelets per cm (MNSC), maximum number of florets per spikelet (MNFS), maximum outer bract length (MOBL), maximum outer bract width (MOBW), maximum middle bract length (MMBL), maximum middle bract width (MMBW), maximum inner bract length (MIBL), maximum inner bract width (MIBW), and maximum calyx length (MCL). All traits were measured in the lab, after removal of flower parts of each individual. Statistical evaluations were performed with the program SPSS 20 (IBM SPSS, 2010) for Windows. The morphometric variables were tested for deviations from a normal distribution using a Kolmogorov-Smirnov test, and then these variables were log transformed. CDA were conducted to give indication of the degree to which the species were distinguishable from each other and to determine which characters contributed to this discrimination. The box-plots showing the medians and interquartile ranges were produced for each significant character for each species.
DNA isolation
Three replicate DNA extractions from leaves of each sample were performed from c. 0.05 g silica gel dried leaf material using the using the DNeasy 96 Plant Kit (Qiagen, UK) and the Mixer Mill MM 300 (Retsch, Haan, Germany) according to the manufacturers’ instructions. DNA quality and quantity were verified using the nanodrop 2000 spectrophotometer (ThermoScientific, Wilmington, USA). Isolated DNA was diluted in nanopure water to produce working stocks of approximately10 ng μl-1.
Genetic/Epigenetic analyses - MSAP procedure
We used a modification of the MSAP protocol to reveal global variability in CG methylation patterns between samples of the different specimens studied [36]. For each individual, 50 ng of DNA was first digested and ligated using 5U of EcoRI and 1U of MspI or HpaII (New England Biolabs), 0.45 μM EcoRI adaptor, 4.5 μM HpaII adaptor and 1U of T4 DNA ligase (Sigma) in 11 μL total volume of 1X T4 DNA ligase buffer (Sigma), 1 μL of 0.5 M NaCl, supplemented with 0.5 μL at 1 mg/ml of BSA for 2 h at 37°C. The enzymes were then inactivated by heating to 75°C for 15 min. Following restriction and adaptor ligation, there followed two successive rounds of PCR amplification. For pre-selective amplification, 0.3 μL of the restriction/ligation products described above were incubated in 12.5 μL volumes containing 1X Biomix (Bioline, London, UK) with 0.05 μL of PreampEcoRI primer and 0.25 μL PreampHpaII/MspI (both primers at 10 lM) (Additional file 4) supplemented with 0.1 μL at 1 mg/ml of BSA. PCR conditions were 2 min at 72°C followed by 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 2 min with a final extension step of 10 min at 72°C. Selective PCRs were then performed using 0.3 μL of pre-selective PCR products and the same reagents as deployed for the pre-selective reactions but using FAM labeled selective primers (Additional file 4) Cycling conditions for selective PCR were as follows: 94°C for 2 min, 13 cycles of 94°C for 30 s, 65°C (decreasing by 0.7°C each cycle) for 30 s, and 72°C for 2 min, followed by 24 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min, ending with 72°C for 10 min. Initially, eight selective primer combinations (Additional file 4) were evaluated for their ability to detect of inter-specific variation and to generate informative and consistent MSAP profiles using two replicated samples from six different populations (data not shown). Two primer combinations (E1/H1 and E1/H3; Additional file 4) were chosen for the comparative selective amplification.
Fluorescently labelled MSAP products were diluted 1:10 in nanopure sterile water and 1 μL was combined with 1 μL of ROX/HiDi mix (50 μL ROX plus 1 ml of HiDi formamide, Applied Biosystems, USA). Samples were heat-denatured at 95°C for 3–5 min and snap-cooled on ice for 2 min. Samples were fractionated on an ABI PRISM 3100 at 3 kV for 22 s and at 15 kV for 45 min.
Data analysis
MSAP profiles were analysed using Genemapper 4.0 software (Applera Corporation, Norwalk, Connecticut, USA). For analysis of the genetic/epigenetic variability between samples revealed using MSAP, reproducible product peaks were scored as present (1) or absent (0) to form a raw data matrix. In order to minimize the occurrence of fragment size homoplasy [69] only fragments with lengths between 100 and 500 bp were considered for the analysis. All monomorphic fragments and any fragments present/absent in all but one individual were considered uninformative and removed from all data sets [70]. Reproducibility was estimated calculating the proportion of dimorphic markers between the replicates of the selected samples [70]. For biological error rates, we compared paired MSAP profiles from two leaves of one plant of six populations using each primer combination. Technical reproducibility of the MSAP technique was assessed through the direct comparison of profiles derived from single DNA extractions from ten representative genotypes.
Analysis of Genetic/Epigenetic Variance
Genetic and epigenetic similarity between tested samples was determined by PCoA [71] based on the MSAP profiles obtained from primer combinations E1/H1 and E1/H3 using GenAlex (v.6.4). Different components of variability were obtained using GenAlex (6.4) software by grouping the samples in two different levels (Populations (i.e. a group of samples) and Regions (i.e. a group of populations). To calculate distances between natural populations, samples from the same natural population restricted with each enzyme were grouped by Populations (this generated two populations from each natural population, one restricted with MspI and one with HpaII) and then at higher level they were grouped into two Regions (all samples restricted with each enzyme). To calculate distances between species, samples from the same taxa restricted with each enzyme were considered and grouped as one Population (this generated two populations from each original species, one restricted with MspI and one with HpaII) and then at higher level they were grouped into two Regions (all samples restricted with each enzyme). We then used AMOVA [72] to evaluate the structure and degree of epigenetic diversity among and between populations, and between species. Pairwise PhiPT [72] comparisons (an analogue of the Fst fixation index, that measures differential connectivity/genetic diversity among populations) between samples restricted with MspI or HpaII was used to infer their overall level of genetic or epigenetic divergence respectively. AMOVA was subsequently calculated using GenAlex (v.6.4) to test the significance of PhiPT between populations and species [73] with the probability of non-differentiation (PhiPT = 0) being estimated over 9,999 permutations. We then calculated genetic diversity estimates (expected heterozygosity, Hj) and the actual genetic diversity for each of the groups above, by using Shannon’s index (ShI) [74]. Finally, pairwise genetic or epigenetic and geographical distance (kilometres) matrices were analysed between populations within species and among species by means of a Mantel test. The level of significance was assigned after 1000 permutation tests, as implemented in Genalex 6 [75].
Abbreviations
- AMOVA:
-
Analysis of Molecular Variance
- CDA:
-
Canonical Discriminant Analysis
- MSL:
-
Maximum spike length
- MNSC:
-
Maximum number of spikelets per cm
- MNFS:
-
Maximum number of florets per spikelet
- MOBL:
-
Maximum outer bract length
- MOBW:
-
Maximum outer bract width
- MMBL:
-
Maximum middle bract length
- MMBW:
-
Maximum middle bract width
- MIBL:
-
Maximum inner bract length
- MIBW:
-
Maximum inner bract width
- MCL:
-
Maximum calyx length
- MSAPs:
-
Methylation Sensitive Amplified Polymorphisms
- PCoA:
-
Principal Coordinates Euclidean Analysis.
References
Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, Leroy CJ, Lonsdorf EV, Allan GJ, DiFazio SP, Potts BM, et al: A framework for community and ecosystem genetics: from genes to ecosystems. Nat Rev Genet. 2006, 7 (7): 510-523. 10.1038/nrg1877.
Johnson MTJ, Stinchcombe JR: An emerging synthesis between community ecology and evolutionary biology. Trends Ecol Evol. 2007, 22 (5): 250-257. 10.1016/j.tree.2007.01.014.
Cubas P, Vincent C, Coen E: An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999, 401 (6749): 157-161. 10.1038/43657.
Manning KTM, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB: A naturally occurring epigenetic mutation in a gene encoding an SBP/box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006, 38 (8): 948-952. 10.1038/ng1841.
Tricker PJ, Gibbings JG, Rodríguez López CM, Hadley P, Wilkinson MJ: Low relative humidity triggers RNA-directed de novo DNA methylation and suppression of genes controlling stomatal development. J Exp Bot. 2012, 63 (10): 3799-3813. 10.1093/jxb/ers076.
Law JA, Jacobsen SE: Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010, 11 (3): 204-220. 10.1038/nrg2719.
Margueron R, Reinberg D: Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010, 11: 285-296. 10.1038/nrg2752.
Rapp RA, Wendel JF: Epigenetics and plant evolution. New Phytol. 2005, 168 (1): 81-91. 10.1111/j.1469-8137.2005.01491.x.
Bossdorf O, Richards CL, Pigliucci M: Epigenetics for ecologists. Ecol Lett. 2008, 11 (2): 106-115.
Richards EJ: Natural epigenetic variation in plant species: a view from the field. Curr Opin Plant Biol. 2011, 14: 204-209. 10.1016/j.pbi.2011.03.009.
Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PC: Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One. 2010, 5 (4): e10326. 10.1371/journal.pone.0010326.
Herrera CM, Bazaga P: Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytol. 2010, 187 (3): 867-876. 10.1111/j.1469-8137.2010.03298.x.
Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A: Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 2010, 185 (4): 1108-1118. 10.1111/j.1469-8137.2009.03121.x.
Erben M: Die gattung Limonium im südwestmediterranen Raum. Mitt Bot Staatssamml München. 1978, 14: 361-631.
Erben M, et al: Limonium Miller. Flora Iberica III Plumbaginaceae (Partim)-Capparaceae. Edited by: Castroviejo S, Aedo C, Cirujano S. Madrid, Real Jardín Botánico: CSIC (Consejo Superior de Investigaciones Científicas); 1993, 2-143.
Kubitzki K: Plumbaginaceae. The Families and Genera of Vascular Plants, Volume 2. Edited by: Kubitzki K, Rohwer JG, Bittrich V. Berlin: Springer Verlag; 1993, 523-530.
Baker HG: Dimorphism and monomorphism in the Plumbaginaceae. 3. Correlation of geographical distribution patterns with dimorphism and monomorphism in Limonium. Ann Bot. 1953, 17 (68): 615-627.
Baker HG: Dimorphism and monomorphism in the Plumbaginaceae. 2. Pollen and stigmata in the genus Limonium. Ann Bot. 1953, 17 (67): 433-445.
Baker HG: The evolution, functioning and breakdown of heteromorphic incompatibility systems. I. The Plumbaginaceae. Evolution. 1966, 20: 349-368. 10.2307/2406635.
D'Amato F: Triploidia e apomissia in Statice oleifolia Scop. var. confusa Godr. Caryologia. 1949, 401: 157-161.
Hjelmqvist HGF: Studies on variation in embryo sac development. Bot Not. 1964, 117: 141-166.
Ingrouille M: The colony structure and taxonomic characterisation of apomitic Limonium (Plumbaginaceae) in the British Isles. PhD thesis. United Kingdom: University of Leicester; 1982.
Palacios C, Rosselló JA, González-Candelas F: Study of the evolutionary relationships among Limonium species (Plumbaginaceae) using nuclear and cytoplasmic molecular markers. Mol Phylogenet Evol. 2000, 14 (2): 232-249. 10.1006/mpev.1999.0690.
Lledó MDCM, Cameron KM, Fay MF, Chase MW: Systematics of Plumbaginaceae based upon cladistics analysis of rbcL sequence data. Syst Bot. 1998, 23: 21-29. 10.2307/2419571.
Lledó MD, Karis PO, Crespo MB, Fay MF, Chase MW: Phylogenetic position and taxonomic status of the genus Aegialitis and subfamilies Staticoideae and Plumbaginoideae (Plumbaginaceae): evidence from plastid DNA sequences and morphology. Plant Syst Evol. 2001, 229 (1–2): 107-124.
Lledó MD, Crespo MB, Fay MF, Chase MW: Molecular phylogenetics of Limonium and related genera (Plumbaginaceae): biogeographical and systematic implications. Am J Bot. 2005, 92 (7): 1189-1198. 10.3732/ajb.92.7.1189.
Santo MDE, Alves HN, Caperta AD, Moreira I: Plantas endémicas do litoral de Portugal Continental. Gestão e Conservação da Flora e da Vegetação de Portugal e da África Lusófona. "In Honorium" do Professor Catedrático Emérito Ilídio Rosário dos Santos Moreira. Edited by: Monteiro A, Gomes da Silva F, Jorge R. 2012, Lisboa: IsaPress, 267-302.
Erben M: Limonium nydeggeri eine neue Art aus Sudwestportugal. Sendtnera. 1999, 6: 103-107.
Commission of the European Communities: Commission decision of 19 July 2006 adopting, pursuant to Council Directive 92/43/EEC, the list of sites of Community importance for the Mediterranean biogeographical region. Off J Eur Union. 2006, L 259: 1-104.
Franco JA: Edição do Autor. Nova Flora de Portugal (Continente e Açores), Volume II, Clethraceae - Compositae. 1984, Lisboa
Rois AS, Teixeira G, Sharbel TF, Fuchs J, Martins S, Espirito-Santo D, Caperta AD: Male fertility versus sterility, cytotype, and DNA quantitative variation in seed production in diploid and tetraploid sea lavenders (Limonium sp., Plumbaginaceae) reveal diversity in reproduction modes. Sex Plant Rep. 2012, 25 (4): 305-318. 10.1007/s00497-012-0199-y.
Baker HG: Dimorphism and monomorphism in the Plumbaginaceae. 1. A survey of the family. Ann Bot. 1948, 12 (47): 207-219.
Dawson HJ, Ingrouille MJ: A biometric survey of Limonium vulgare Miller and L. humile Miller in the British Isles. Watsonia. 1995, 20: 239-254.
Kremer A, Caron H, Cavers S, Colpaert N, Gheysen G, Gribel R, Lemes M, Lowe AJ, Margis R, Navarro C, et al: Monitoring genetic diversity in tropical trees with multilocus dominant markers. Heredity. 2005, 95 (4): 274-280. 10.1038/sj.hdy.6800738.
Garcia-Verdugo C, Forrest AD, Fay MF, Vargas P: The relevance of gene flow in metapopulation dynamics of an oceanic island endemic. Olea europaea subsp. guanchica. Evolution. 2010, 64 (12): 3525-3536. 10.1111/j.1558-5646.2010.01091.x.
Reyna-Lopez GE, Simpson J, Ruiz-Herrera J: Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol Gen Genet. 1997, 253 (6): 703-710. 10.1007/s004380050374.
Paun O, Schönswetter P: Amplified fragment length polymorphism: an invaluable fingerprinting technique for genomic, transcriptomic, and epigenetic studies. Plant DNA Fingerprinting and Barcoding – Methods and Protocols, Volume 862. Edited by: Sucher NJ, Hennell JR, Carles MC. 2012, New York: Humana Press, 75-87.
Morán P, Pérez-Figueroa A: Methylation changes associated with early maturation stages in the Atlantic salmon. BMC Genet. 2011, 12: 86. 10.1186/1471-2156-12-86.
Mathieu O, Reinders J, Čaikovski M, Smathajitt C, Paszkowski J: Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007, 130: 851-862. 10.1016/j.cell.2007.07.007.
Tricker P, Rodríguez López CM, Gibbings G, Hadley P, Wilkinson MJ: Transgenerational, dynamic methylation of stomata genes in response to low relative humidity. Int J Mol Sci. 2013, 14 (4): 6674-6689. 10.3390/ijms14046674.
Ingrouille MJ: A taxometric analysis of Limonium (Plumbaginaceae) in Western-Europe. Plant Syst Evol. 1984, 147 (1–2): 103-118.
Ingrouille MJ, Stace CA: Pattern of variation of agamospermous Limonium (Plumbaginaceae) in the British-Isles. Nordic J Bot. 1985, 5 (2): 113-125.
Cowan R, Ingrouille MJ, Lledó MD: The taxonomic treatment of agamosperms in the genus Limonium Mill. (Plumbaginaceae). Folia Geobot. 1998, 33 (3): 353-366. 10.1007/BF03216212.
Asker SE: Jerling: Apomixis in Plants. Boca Raton: CRC Press; 1992
Paun O, Greilhuber J, Temsch EM, Horandl E: Patterns, sources and ecological implications of clonal diversity in apomictic Ranunculus carpaticola (Ranunculus auricomus complex, Ranunculaceae). Mol Ecol. 2006, 15 (4): 897-910. 10.1111/j.1365-294X.2006.02800.x.
Carman JG: Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc. 1997, 61 (1): 51-94. 10.1111/j.1095-8312.1997.tb01778.x.
Carman JG: The gene effect: Genome collision and apomixis. The Flowering of Apomixis: From Mechanisms to Genetic Engineering. Edited by: Savidan Y, Carman JG, Dresselhaus T. 2001, Mexico: CIMMYT, 95-110.
Carman JG: Do duplicate genes cause apomixis?. Apomixis: Evolution, Mechanisms and Perspectives. Edited by: Hörandl E, Grossniklaus U, Van-Dijk P, Sharbel T. Ruggell, Liechtenstein: Gantner Verlag [Regnum Vegetabile 147]; 2007, 169-194.
Wilkinson MJ, Stace CA: A new taxonomic treatment of the Festuca ovina L. aggregate (Poaceae) in the British Isles. Bot Jour Linn Soc. 1991, 106: 347-397. 10.1111/j.1095-8339.1991.tb02298.x.
Bretagnolle F, Thompson JD: Phenotypic plasticity in sympatric diploid and autotetraploid Dactylis glomerata. Int J Plant Sci. 2001, 162 (2): 309-316. 10.1086/319572.
Kolar F, Fer T, Stech M, Travnicek P, Duskova E, Schonswetter P, Suda J: Bringing together evolution on serpentine and polyploidy: spatiotemporal history of the diploid-tetraploid complex of Knautia arvensis (Dipsacaceae). PLoS One. 2012, 7 (7): e39988. 10.1371/journal.pone.0039988.
Nylehn J, Hamre E, Nordal I: Facultative apomixis and hybridization in arctic Potentilla section Niveae (Rosaceae) from Svalbard. Bot Jour Linn Soc. 2003, 142: 373-381. 10.1046/j.1095-8339.2003.00190.x.
Ingrouille MJ, Stace CA: The Limonium binervosum aggregate (Plumbaginaceae) in the British Isles. Bot Jour Linn Soc. 1986, 92 (3): 177-217. 10.1111/j.1095-8339.1986.tb01428.x.
Rodríguez S, Palop ML, Palacios C, González-Candelas F: Molecular and morphological differentiation in Limonium dufourii (Plumbaginaceae), an endangered Mediterranean plant. Cons Genet. 2003, 4 (3): 383-391. 10.1023/A:1024085517637.
te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M, Pysek P: The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot. 2012, 109 (1): 19-45. 10.1093/aob/mcr277.
Palacios C, González-Candelas F: Analysis of population genetic structure and variability using RAPD markers in the endemic and endangered Limonium dufourii (Plumbaginaceae). Mol Ecol. 1997, 6 (12): 1107-1121. 10.1046/j.1365-294X.1997.00283.x.
Palacios C, Kresovich S, González-Candelas F: A population genetic study of the endangered plant species Limonium dufourii (Plumbaginaceae) based on amplified fragment length polymorphism (AFLP). Mol Ecol. 1999, 8 (4): 645-657. 10.1046/j.1365-294X.1999.t01-1-00597.x.
Palop-Esteban M, Segarra-Moragues JG, González-Candelas F: Historical and biological determinants of genetic diversity in the highly endemic triploid sea lavender Limonium dufourii (Plumbaginaceae). Mol Ecol. 2007, 16 (18): 3814-3827. 10.1111/j.1365-294X.2007.03449.x.
Suárez-García C, de Paz JP, Febles R, Caujapé-Castells J: Genetic diversity and floral dimorphism in Limonium dendroides (Plumbaginaceae), a woody Canarian species on the way of extinction. Plant Syst Evol. 2009, 280 (1–2): 105-117.
Kalisz S, Vogler DW, Hanley KM: Context-dependent autonomous self-fertilization yields reproductive assurance and mixed mating. Nature. 2004, 430 (7002): 884-887. 10.1038/nature02776.
Holliday R: The biological significance of meiosis. Symp Soc Exp Biol. 1984, 38: 381-394.
Adams KL, Cronn R, Percifield R, Wendel JF: Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. PNAS. 2003, 100 (8): 4649-4654. 10.1073/pnas.0630618100.
Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF: Evolutionary genetics of genome merger and doubling in plants. Ann Rev Genet. 2008, 42: 443-461. 10.1146/annurev.genet.42.110807.091524.
Fortune PM, Schierenbeck KA, Ainouche AK, Jacquemin J, Wendel JF, Ainouche ML: Evolutionary dynamics of Waxy and the origin of hexaploid Spartina species (Poaceae). Mol Phyl Evol. 2007, 43 (3): 1040-1055. 10.1016/j.ympev.2006.11.018.
Cortinhas AL: MSc thesis. Estudio de los taxa afines a Limonium vulgare Miller de las marismas de Portugal Continental. Spain: Universidad de Valencia: 2012.
Richards AJ: Plant Breeding Systems 2 nd edition. London: Chapman and Hall; 1997.
Erben M: Karyotype differentiation and its consequences in Mediterranean Limonium. Webbia. 1979, 34: 409-417. 10.1080/00837792.1979.10670178.
Hutchison DW, Templeton AR: Correlation of pairwise genetic and geographic distance measures: Inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999, 53: 1898-1914. 10.2307/2640449.
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I: Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol. 2002, 11: 139-151. 10.1046/j.0962-1083.2001.01415.x.
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P: How to track and assess genotyping errors in population genetics studies. Mol Ecol. 2004, 13 (11): 3261-3273. 10.1111/j.1365-294X.2004.02346.x.
Gower JC: Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966, 53 (3–4): 325-338.
Excoffier L, Smouse PE, Quattro JM: Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992, 131 (2): 479-491.
Michalakis Y, Excoffier L: A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics. 1996, 142: 1061-1064.
Abbott RJ, Ireland HE, Rogers HJ: Population decline despite high genetic diversity in the new allopolyploid species Senecio cambrensis (Asteraceae). Mol Ecol. 2007, 16 (5): 1023-1033. 10.1111/j.1365-294X.2007.03169.x.
Peakall ROD, Smouse PE: Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006, 6 (1): 288-295. 10.1111/j.1471-8286.2005.01155.x.
Acknowledgements
The authors wish to thank Ana Paula Paes (CBAA) for help in field work and Wanda Viegas (CBAA) for critical insights. We thank the Instituto da Conservação da Natureza e Biodiversidade (ICNB) for providing permissions for collecting samples. ADC (Researcher, CBAA/ISA) and ASR (SFRH/BD/62542/2009 grant) were supported by FUNDAÇÃO PARA A CIÊNCIA E TECNOLOGIA, FCT, Portugal). The project was funded by PEST-OE/AGR/UI0240/2011.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
ADC designed and coordinated the study. ADC and ASR performed plant sampling. ASR and CMRL contributed to the experimental design and performed the molecular genetic studies by MSAPs. AC and ME realized the morphological measurements. ASR, CMRL and AC processed the raw data and carried out the statistical analysis. CMRL and ADC drafted the manuscript. DES and MJW critically revised the manuscript. All authors read and approved the manuscript.
Ana Sofia Róis, Carlos M Rodríguez López contributed equally to this work.
Electronic supplementary material
12870_2013_1428_MOESM1_ESM.png
Additional file 1: Mean values of morphometric characters in diploid and tetraploid Limonium species. Diploid L. lanceolatum, L. nydeggeri, L. ovalifolium and tetraploid L. dodartii, L. multiflorum and L. vulgare species are considered. (PNG 78 KB)
12870_2013_1428_MOESM2_ESM.png
Additional file 2: Principal Coordinate Analysis (PCoA) representing genetic and epigenetic variability in diploid and tetraploid Limonium species. PCoA was based on presence/absence scores of 347 polymorphic loci obtained from MSAP profiles using isoschizomers MspI (methylation insensitive - red symbols in A and B) or HpaII (methylation sensitive - blue symbols in A and C) as frequent cutters and amplified with primers (E1/H1). The first two coordinates were extracted and plotted against each other. Percentage of the variability shown by each coordinate is indicated between parentheses. Diploid species are represented by solid symbols (L. lanceolatum, triangles; L. nydeggeri, rhomboids; L. ovalifolium, rectangles) and tetraploid species are represented by empty symbols (L. dodartii, triangles; L. multiflorum, rectangles; L. vulgare, circles). (PNG 186 KB)
12870_2013_1428_MOESM3_ESM.png
Additional file 3: Estimated genetic (GD) and epigenetic (EpiDG) distances between and within diploid and tetraploid Limonium species. Diploid (L. lanceolatum, L. nydeggeri, L. ovalifolium) and tetraploid (L. dodartii, L. multiflorum and L. vulgare) species are considered. (PNG 109 KB)
12870_2013_1428_MOESM4_ESM.png
Additional file 4: Oligonucleotides used for MSAP analysis. Selective nucleotides are indicated as + XYZ in the primer code column. Enzyme column indicates the restriction enzyme site associated with each primer. * FAM labeled selective primers. (PNG 46 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Róis, A.S., Rodríguez López, C.M., Cortinhas, A. et al. Epigenetic rather than genetic factors may explain phenotypic divergence between coastal populations of diploid and tetraploid Limonium spp. (Plumbaginaceae) in Portugal. BMC Plant Biol 13, 205 (2013). https://doi.org/10.1186/1471-2229-13-205
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-13-205