Abstract
Background
The Arabidopsis microRNA156 (miR156) regulates 11 members of the SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) family by base pairing to complementary target mRNAs. Each SPL gene further regulates a set of other genes; thus, miR156 controls numerous genes through a complex gene regulation network. Increased axillary branching occurs in transgenic Arabidopsis overexpressing miR156b, similar to that observed in loss-of-function max3 and max4 mutants with lesions in carotenoid cleavage dioxygenases. Arabidopsis miR156b was found to enhance carotenoid levels and reproductive shoot branching when expressed in Brassica napus, suggesting a link between miR156b expression and carotenoid metabolism. However, details of the miR156 regulatory network of SPL genes related to carotenoid metabolism are not known.
Results
In this study, an Arabidopsis T-DNA enhancer mutant, sk156, was identified due to its altered branching and trichome morphology and increased seed carotenoid levels compared to wild type (WT) ecovar Columbia. Enhanced miR156b expression due to the 35S enhancers present on the T-DNA insert was responsible for these phenotypes. Constitutive and leaf primodium-specific expression of a miR156-insensitive (mutated) SPL15 (SPL15m) largely restored WT seed carotenoid levels and plant morphology when expressed in sk156. The Arabidopsis native miR156-sensitive SPL15 (SPL15n) and SPL15m driven by a native SPL15 promoter did not restore the WT phenotype in sk156. Our findings suggest that SPL15 function is somewhat redundant with other SPL family members, which collectively affect plant phenotypes. Moreover, substantially decreased miR156b transcript levels in sk156 expressing SPL15m, together with the presence of multiple repeats of SPL-binding GTAC core sequence close to the miR156b transcription start site, suggested feedback regulation of miR156b expression by SPL15. This was supported by the demonstration of specific in vitro interaction between DNA-binding SBP domain of SPL15 and the proximal promoter sequence of miR156b.
Conclusions
Enhanced miR156b expression in sk156 leads to the mutant phenotype including carotenoid levels in the seed through suppression of SPL15 and other SPL target genes. Moreover, SPL15 has a regulatory role not only for downstream components, but also for its own upstream regulator miR156b.
Similar content being viewed by others
Background
The Arabidopsis miR156 family has eight members and is highly conserved in the plant kingdom. It has been identified in 45 plant species [1]. MiR156 is known to repress SPL (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE) genes [2–4], which are plant-specific transcription factors containing the SBP (SQUAMOSA promoter binding protein) box [5]. In Arabidopsis, 11 SPL genes are targeted by miR156 [2, 4, 6, 7], and in silico full genome analysis showed that no other genes in Arabidopsis have the segment complimentary to miR156 [2]. Some details of the relationship between the miR156 regulatory network downstream of SPL genes and flowering enhancement and flavonoid metabolism have been revealed [8–10]. But this is clearly an area of research that still needs strong attention.
Diverse and redundant roles of some individual SPL genes in plant morphology and development have been reported. The SPL genes targeted by miR156 can be grouped into four major clades: SPL3/SPL4/SPL5, SPL2/SPL10/SPL11, SPL9/SPL15, and SPL6/SPL13[7]. SPL3, SPL4 and SPL5 exhibit partially redundant effects on plant juvenile-to-adult transition [4, 11, 12]. SPL9 and SPL3 directly activate MADS box genes that promote flowering [8]. In addition, SPL9 and SPL15 interchangeably control shoot maturation and leaf initiation [13]. SPL10 and SPL9 expression in leaf primordia modulated by miR156 affects initiation of new leaves at the shoot apical meristem [14]. SPL2, SPL10 and SPL11 were each able to control leaf lamina shape in association with shoot maturation in the reproductive phase [15]. Moreover, SPL genes (represented by SPL9) regulate trichome development via direct interaction with miR172 [9] and the MYB transcription factor genes TRICHOMELESS1 (TCL1) and TRIPTYCHON (TRY) [16].
Increased axillary branching occurs in transgenic Arabidopsis lines expressing miR156b under the control of a cauliflower mosaic virus 35S promoter (CaMV35S) [6], similar to that observed in loss-of-function max3 and max4 mutants with lesions in carotenoid cleavage dioxygenases, CCD7 and CCD8, respectively [17–19]. These max mutations cause defective biosynthesis of strigolactones; a group of carotenoid-derived hormones that inhibit shoot branching [20, 21]. The morphological similarities between these transgenic and mutant lines suggest a link between miR156b expression and carotenoid metabolism. This was partially confirmed when Arabidopsis miR156b was found to enhance carotenoid levels and reproductive shoot branching when expressed in Brassica napus[22].
In this study, we report on a new Arabidopsis activation tagged mutant, sk156, with strongly enhanced expression of miR156b. MiR156b-induced SPL15 suppression was partially responsible for the increased seed carotenoid abundance and altered plant morphology observed in sk156. In addition, we highlight a new SPL15 feedback loop which controls expression of miR156b, likely via a physical interaction between the SPL15 SBP domain and the promoter of miR156b.
Results
Phenotypes of the sk156mutant
The sk156 mutant was selected from an Arabidopsis activation-tagged mutant population that was developed using a T-DNA construct containing four CaMV35S enhancers [23]. Compared to the parental wild type (WT) Col-4, sk156 exhibited the following morphological changes: increased numbers of rosette leaves which were slightly pale, smaller and rounder, increased reproductive branching, ectopic expression of trichomes on flower sepals and shoot tips, delayed bolting, severely stunted cauline stems, and decreased flower and silique sizes (Figure 1).
Morphological phenotype of the Arabidopsis sk156 mutant. (A) 30-d-old rosette plants. (B) 8-week-old mature plants. (C) 30-d-old plant leaves. WT on top, sk156 on bottom. (D) Flowers, showing trichome-bearing sk156 sepals (right). (E) Siliques at 14 days post-anthesis. (F) Trichome enhancement on cauline stems and leaves of sk156. (G) Enhanced rosette leaf and branch numbers (insert defines branch order). Data were the mean of thirty 8-week-old plants. MB, main stem; RL, rosette leaves. A Duncan's multiple range test showed significant differences of the means (± standard deviation) between mutant lines and WT at p < 0.01 (**) and p < 0.05 (*).
For comparison, we examined the phenotypes of max3-9[17] and max4-1[19] mutants, which had been confirmed previously to contain defective CCD7 and CCD8 genes, respectively (Additional file 1). In our hands, these max mutants showed increased secondary and tertiary branches (Figure 1G), which was consistent with an earlier report that max mutants showed increased inflorescence number compared to WT plants [24]. However the cauline stems of these two max mutants were not significantly stunted as in sk156. The branching pattern of sk156 was also somewhat distinct compared with max mutants; sk156 had more quaternary and quinary branches and the max mutants mainly had increased secondary and tertiary branches.
The abundance of five major carotenoid compounds was examined in the mature seeds and leaves of sk156 and WT plants. Levels of lutein, β-carotene, violaxanthin, and zeaxanthin were 2.3-, 11.7, 5.5-, and 1.8- fold higher in sk156 seeds than in WT seeds (Figure 2A and B). Slight decreases (but not statistically significant) in the levels of major carotenoids in leaves were found in sk156 compared to WT (Figure 2C and D). Cryptoxanthin was undetectable in either tissue of the mutant, but present at 0.62 μg g-1 FW (fresh weight) in WT seeds and 4-fold higher in WT leaves.
Carotenoid levels in sk156 mutant. (A) and (B) Carotenoid levels in mature seeds. (C) and (D) Carotenoid levels in leaves of 30-d-old plants. A Duncan's multiple range test showed significant differences of the means (±standard deviation) between mutant lines and WT at p < 0.01 (**) and p < 0.05 (*).
Molecular analysis of sk156
Segregation analysis of heterozygous sk156 mutant offspring revealed that the ratio of resistant-to-sensitive seedlings (271:109, respectively) was well within the expected 3:1 ratio (χ2, p = 0.05), indicating that there was a single T-DNA insertion site. This was confirmed by Southern blot analysis (Figure 3A). Sequencing of the T-DNA-flanking regions indicated that the T-DNA was located in AT4G30980 (Figure 3B), which encodes a basic helix-loop-helix (bHLH) family protein bHLH069 [25, 26]. CaMV 35S enhancers in the T-DNA were within close proximity to five other gene loci: AT4G30975 (unknown RNA gene), AT4G30972 (miR156b), AT4G30970 (unknown protein) and AT4g30960 (SOS2-like protein kinase PKS4).
T-DNA insertion site and affected genes in sk156 mutant. (A) Southern blot analysis of sk156 showing a single T-DNA insertion. Lane 1, digested with BamHI. Lane 2, digested with HindIII. (B) Schematic of T-DNA inserted in the third intron of bHLH069 shown by ‘∇’ (empty triangle). Boxes, exons; Lines, introns and intergenic regions. Thick grey arrows show promoter regions and their transcription direction. (C) Transcript levels for miR156b and unknown RNA gene (AT4G30975) in sk156 relative to WT (set at 1) using qPCR. (D) Transcript levels for bHLH069 in sk156 relative to WT (set at 1) using qPCR. A Duncan's multiple range test in (C) and (D) showed significant differences of the means (± standard deviation) between sk156 and WT Col-4 at p < 0.01 (**) and p < 0.05 (*).
The sk156 phenotypes were dominant when crossed with WT, suggesting that the altered phenotypes in sk156 were tightly linked to enhanced expression of gene(s) adjacent to the T-DNA, rather than gene disruption. Thus, transcript levels of genes close to the T-DNA insertion site were compared in the mutant and WT plants using quantitative RT-PCR. Transcript levels of AT4G30972 (miR156b) and the unknown RNA gene AT4G30975 were increased 91.4-fold (± 25.8) and 95.8-fold (±18.6), respectively, in sk156 (Figure 3C), while the T-DNA-disrupted AT4g30980 (bHLH069) was repressed 11.1-fold (Figure 3D). No significant changes in transcript levels were detected for the other adjacent genes (data not shown). This indicated that the four CaVM 35S enhancers inserted into bHLH069 caused the activation of miR156b and the unknown RNA gene.
To determine which of the three T-DNA affected genes was responsible for the altered phenotypes in sk156, including enhanced seed carotenoid levels, four types of plants were obtained and examined (Table 1). These included a bHLH069 knockout mutant S468 (SALK_032468) (Figure 4A), transgenic sk156 over-expressing a 35S:bHLH069 cDNA (Figure 4B) transgenic WT carrying 35S:AT4G30975 cassette (A975, the unknown RNA gene) (Figure 4C and D), and transgenic Arabidopsis WT carrying a 35S:miR156b cassette (T156b) (Figure 4E and F) [12]. Transcript analysis showed that expression of bHLH069 was defective in both S468 and sk156 but enhanced in sk156 over-expressing bHLH096 (Figure 4A). Morphology was indistinguishable between sk156 and sk156 over-expressing bHLH096 (Figure 4B) and between the SALK mutant and WT (data not shown). Additionally, over-expression of the unknown RNA cDNA in WT (line A975) (Figure 4C) did not produce the sk156 mutant phenotype (Figure 4D). However, the 35S promoter-driven miR156b overexpression resulted in morphological characteristics in line T156b similar to those of sk156 (Figure 4F).
Involvement of miR156b , bHLH069 , and the unknown RNA AT4G30975 in the phenotypes of the sk156 mutant. (A) Transcript levels of bHLH069 in WT (set at 1), sk156, mutant line S468 (SALK_032468), and in sk156 transformants complemented with a 35S:bHLH069 construct relative to WT (set at 1). Insert, schematic of T-DNA insertion site (triangle) in bHLH069 (AT4G30980) in S468 shows above. Exons (black boxes), untranslated regions (light grey boxes), promoter direction (dark arrow). (B) Morphology of 45-d-old transgenic sk156 complemented with 35S:bHLH069. Sk156, and WT plants are also shown. (C) Transcript levels of unknown RNA gene AT4G30975 in WT (set at 1), sk156, and 35S:AT4G30975cDNA-complemented sk156 (A975) plants. Schematic of AT4G30975 expression construct shows above. (D) Morphology of 35-d-old transgenic plant A975 compared to sk156. (E) Transcript levels of miR156 in WT, sk156, and 35S:miR156b-complemented sk156 (T156b) plants. Schematic of 35S:miR156b expression construct shows above. (F) Morphology of 40-d-old transgenic plants from sk156 and 35S:miR156b-over-expressing T156b lines compared to WT. (G) and (H) Seed carotenoid levels in sk156, T156b, A975, S468, 35S:bHLH069/sk156, and WT plants. A Duncan's multiple range test showed significant differences of the means (± standard deviation) compared to WT at p < 0.05. Means containing the same letter for the same compounds are not significantly different.
Carotenoid analysis further confirmed the role of enhanced miR156b expression in generating the sk156 phenotypes (Figure 4G and H). No significant differences in seed carotenoid abundance were found between the defective bHLH069 mutant S468 and WT (p ≤ 0.05). Complementation of sk156 with bHLH069 did not restore sk156 seed carotenoid levels back to WT levels, nor did over-expression of the unknown RNA in WT result in enhanced seed carotenoid levels. Only in line T156b overexpressing miR156b were seed carotenoid levels raised to levels found in sk156 seeds (Figure 4G and H). These data clearly indicate a role for miR156b in the phenotypes of sk156.
Due to similar enhanced transcript levels of miR156b and AT4G30975 in the sk156 mutant, yet unchanged morphology and seed carotenoid accumulation in the AT4G30975 over-expression lines (line A795, Table 1), we examined the relationship between miR156b transcripts and AT4G30975 cDNA. These two transcripts are separated by a 17 bp intergenic fragment (TAIR, version 9). Sequencing of cDNA obtained from the leaves of 4-week-old WT plants revealed a miR156b transcript which included both the 17 bp fragment and the full segment of the AT4G30975 transcript (sequencing data not shown). These data show that enhanced expression of miR156 is the reason for the phenotypic changes in the sk156 mutant line, regardless of AT4G30975 co-transcription.
Suppression of SPL15 is involved in the sk156phenotype
Arabidopsis Columbia has 11 SPL genes (two loci for SPL13) that are regulated by miR156 [7, 9]. To determine which SPL geneswere most likely involved in altered carotenoid profiles of sk156 seeds, mutant lines with confirmed T-DNA insertion knockouts for all miR156-target SPL genes (except SPL5 where mutants were unavailable) were obtained (Additional file 2). The mutant lines were compared for morphology and carotenoid differences with their corresponding ecotype Columbia and Wassilevskija controls. While levels of carotenoids were modestly increased in three spl15 mutant lines (Figure 5A insert), they were less affected in the remaining spl mutants (Figure 5A). Even in the spl15 mutant lines, the enhanced total carotenoid levels (21.5 ± 1.7μg/g.FW) (Figure 5 insert) were not as high as the sum of each individual carotenoid compound observed in sk156 (37.8 ± 5.5μg/g.FW) (Figure 6C and D).
Involvement of SPL15 in phenotypic alteration of sk156 . (A) Carotenoid levels in SPL loss-of-function mutants specifying nine SPL genes, including SPL15. SPL15 SALK lines in insert show details on specific carotenoid changes. (B) SPL15 transcript levels in sk156, SPL15-related Salk lines, and T156b relative to WT (Col, set at 1). Insert shows schematic diagrams of T-DNA insertion sites ‘∇’ in SPL15 SALK mutants CS2117, CS6815 and S8712.Duncan's multiple range test showed significant differences (*) of the means (± standard deviation) between sk156 and WT (Col or WS) at p < 0.05.
Expression of mutated SPL15m in sk156 results restores a WT phenotype and down regulates miR156b transcription. (A) Complementarity of miR156 with SPL15 sequences. Arrows show three cleavage sites (one used 10 out of 13 times) in SPL15 mRNA due to interaction with miR156b. SPL15m shows 11 nucleotides mutated in DNA sequence but unchanged in amino acid sequence. (B) SPL15 transcript levels in independent transgenic sk156 lines carrying 35S:SPL15m compared to WT (set at 1) and sk156. Error bars are standard deviations of the means. (C) and (D) Restoration of WT-like carotenoid levels in dry seeds of transgenic sk156 expressing SPL15m. Cryptoxanthin was undetectable in the mutant. (E) Restoration of WT-like plant growth in 35-d-old plants of sk156 expressing SPL15m. (F) Distinctive elongated leaf morphology of SPL15m-transformed plants compared to WT plants. (G) miR156b transcript levels in different transgenic sk156 lines expressing SPL15m compared to WT (set at 1) and sk156. A Duncan's multiple range test was conducted to separate significantly different means. Panels B, G, ± standard deviation; Panels C, D, ± standard error; p < 0.01 (**) and p < 0.05 (*).
The three different SPL15 “knockout” mutants, CS852117, CS856815 and SALK_138712 (Figure 5A insert) all exhibited reduced SPL15 transcript levels similar to levels found in the sk156 mutant and the miR156b over-expression line T156b (Figure 5B; Table 1). However, other spl mutants had normal cauline stem lengths (data not shown), while spl15 mutants had slightly stunted stems (Additional file 3) as reported previously [13]. Assays using rapid amplification of 5' complementary DNA ends (5'-RACE) indicated that three cleavage sites exist within the region of the native SPL15 transcript that is complementary to the miR156 mature sequence, with one site being used more frequently than the other two (Figure 6A). Collectively, these results suggest that SPL15 is a direct target of miR156 and that it plays a role in the morphology and seed carotenoid phenotypes found in sk156 mutant line.
Expression of miR156-insensitive SPL15m affects the phenotype of sk156
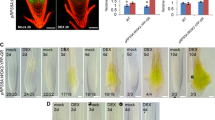
Since spl15 mutants had several phenotypes similar to those of sk156, two different SPL15-complemented sk156 lines were developed to determine whether the WT phenotype could be restored in a miR156b-enhanced sk156 background (Table 1). The first transgenic sk156 carried a 35S:SPL15n (native SPL15) cassette to avoid any endogenous transcriptional regulation which might be present if using a native SPL15 promoter (Figure 6A). The second carried 35S: (mutated SPL15), which contains 11 mutated sites within the SPL15 segment complementary to mature miR156 so that SPL15m becomes insensitive to miR156 regulation (Figure 6A). PCR analysis using gene-specific primers confirmed both pSKI015 T-DNA and either SPL15n or SPL15m within these transgenic lines (Additional file 4). Sequencing of these PCR products confirmed that all SPL15m-expressing lines had a SPL15-mutated segment (Figure 6A) as expected. SPL15 transcript levels (sum of SPL15m and endogenous SPL15) within the 35S:SPL15m+ lines were increased by 86.2-fold to 617.4-fold compared to WT (Figure 6B). No significant differences were found in the amount of lutein, vialoxanthin, zeaxanthin and β-carotene in seeds between these 35S:SPL15m-transformed plants and wild type control plants (Figure 6C and D). The transgenic plants showed a largely restored WT morphology in leaf and branch numbers, bolting time, cauline stem length, flower and silique size, and trichome number in reproductive shoots and tissues (Figure 6E). Only leaf morphology was different between transgenic and non-transgenic lines (Figure 6F). This contrasted with miR156-controlled 35S:SPL15n+ lines (Table 1), which displayed sk156 phenotypes.
Since constitutive expression of SPL15m predominantly restored sk156 phenotypes, native SPL15n and mutated SPL15m under the control of the young leaf primordia promoter from the ASYMMETRIC LEAVES 1 (AS1) gene [27] were used to generate two additional complemented sets of sk156 lines. This was to find out whether SPL15 functions at the leaf primordia within the shoot apex as occurs with certain SPL genes controlling plastochron length [14]. PCR analyses using promoter-specific and gene-specific primers (Additional file 5, see materials and methods) confirmed the presence of the transgene in these transgenic plants (Additional file 6). SPL15m transcript levels were increased in AS1 promoter-driven SPL15m transgenic sk156 lines, but not in AS1 promoter-SPL15n lines, compared to WT and sk156 mutants (Figure 7A). As with SPL15m expression directed by the 35S promoter, expression of SPL15m under the control of the AS1 promoter in the sk156 background restored WT morphology at different growth stages, whereas AS1:SPL15n did not (Figure 7B, C, and D). Moreover, the levels of seed carotenoids in these ASI:SPL15m plants were not significantly different from those in WT (p > 0.05), while seed carotenoid abundance was significantly higher in plants carrying the AS1:SPL15n cassette (p > 0.05) (Figure 7E and F). These changes in the phenotype of transgenic sk156 plants expressing the miR156-insensitive AS1:SPL15m suggested that SPL15m could function independently of miR156b and that expression of SPL15 is effective at the leaf primordia within the shoot apex.
Leaf primodium-dependent miR156-insensitive SPL15m restores WT phenotypes when expressed in sk156 . The sk156 background was transformed with native miR156-sensitive SPL15n or miR156-insensitive SPL15m expression cassettes each controlled by either an AS1 or 35S promoter. (A) SPL15 transcript levels by qPCR (± standard deviation) in transgenic sk156 plants complemented with AS1:SPL15m, AS1:SPL15n, 35S:SPL15n or SPL15:SPL15m compared to WT (set at 1) and sk156 (p < 0.05). (B) and (C) Morphology at bolting (32 d) and maturity (50 d) for plant expressing SPL15n or SPL15m under the control of AS1 promoter compared to sk156. (D) Morphology of 42 d flowering lines for sk156 plant carrying 35S:SPL15n or SPL15:SPL15m compared to sk156. (E) and (F) Carotenoid levels (± standard error) in seeds of sk156 complemented with AS1:SPL15m, AS1:SPL15n , 35S:SPL15n or SPL15:SPL15m compared to those of WT and sk156. Panels A, E, F: A Duncan's multiple range test was conducted to separate significantly different means for more than 10 independent transgenic lines (each measured with triplicated seed batches) relative to WT plants at p < 0.05 (*).
Since the native SPL15n appeared to be ineffective at restoring the WT phenotype in the enhanced miR156b environment of sk156, another set of complemented sk156 lines was generated to express the miR156-insensitive SPL15m under the control of the native SPL15 promoter (Table 1). Morphological traits and carotenoid levels remained unchanged for these transgenic SPL15:SPL15m plants compared to uncomplemented sk156, except that the leaf shape was similarly elongated as were 35S:SPL15m plants compared to WT plants (Figure 6F). These data suggested that overwhelming miR156b levels within the sk156 mutant negatively regulated the expression of the SPL15n under its native promoter and prevented restoration of WT seed carotenoid abundance and plant morphology. Moreover, the inability of SPL15:SPL15m plants to restore most of the WT phenotype as did 35S:SPL15m+ lines suggested that the enhanced seed carotenoid level and morphological traits observed in the sk156 mutant were the collective result of miR156 suppression of at least some other SPL genes rather than SPL15 alone and that SPL15 functions redundantly with some other SPL gene products.
Transcription of miR156b is affected by over-expression of SPL15m
Wu and Poethig [12] reported earlier that expression of miR156-insensitive SPL3 under the control of the CaMV35S promoter causes reduced levels of miR156 transcripts in Arabidopsis. To investigate the ability of SPL15m to affect expression of its regulatory miR156 genes, we examined miR156b transcript levels in the sk156 mutant and in complemented sk156 carrying the 35S:SPL15m cassette (Table 1). Primers for premature miR156b rather than mature miR156 sequence were used so that measurement of transcripts originating from other miR156 genes would be excluded. In 30-day-old plants, miR156b premature transcript levels were reduced from 82.2-fold above WT levels in sk156 leaves down to a mean of 7 ± 5.5 fold above WT in the leaves of these complemented lines, with a minimum of 1.4-fold in line 7 and a maximum of 19.6-fold in line 9 (Figure 6G). Thus, miR156b transcription was depressed substantially such that ~50% of the complemented lines had WT levels. A reduction of miR156b transcript level concurrent with an increase in SPL15m transcripts suggests that SPL15 may have two regulatory functions; one controlling feedback regulation of its cognate regulator miR156 and one controlling downstream genes, such as miR172, APETALA2-LIKE (AP2-like) transcription factors TOE1, and TOE2, in the miR156 controlled gene network [9].
Physical interaction between the SPL15 DNA binding domain and GCAT motifs in the miR156bpromoter
To investigate whether SPL15 interacts directly with the miR156b promoter to affect transcription, we identified putative SPL15 binding sites in the miR156b promoter based on the consensus sequence for SBP binding domain (Figure 8A). To define the consensus sequence, we compared cis elements previously identified as SBP box interacting sequences in Chlamydomonas reinhardtii[28, 29], Antirrhinum majus[30] and A. thaliana[5, 11, 31, 32] using WebLogo [33]. The consensus SBP domain binding sequence was determined to be 5’-NNGTACR-3’, where frequently N = C and R = A (Figure 8A). This consensus sequence was used to search for putative SPL binding sites in the promoters of miR156 genes. Putative SPL binding sites with a GTAC core sequence were repeatedly present in the miR156b promoter [-1 to -1500], according to the gene transcript start site revealed by Schwab [34]. Of these, a region containing three repeats of the core sequence appeared between -200 bp to -220 bp (Figure 8B) suggesting that miR156b expression could be directly controlled by SPL15. This was tested using a His-tagged SPL15 recombinant SBP DNA-binding domain (Figure 8C and D) in an electrophoretic mobility shift assay with a labeled DNA fragment (46-mer) containing a motif from -200 bp to -220 bp from the miR156b promoter. The recombinant SBP domain of SPL15n specifically bound to this region of the miR156b promoter to yield a less mobile promoter fragment (Figure 8E), and competition with 100-fold unlabeled miR156b promoter DNA was required to displace the SBP domain. This result suggested that a direct physical interaction can occur between the SPL15 DNA binding domain and the miR156b promoter through the GTAC motif.
Interaction of the miR156 SBP-binding motif (GTAC core sequence) with the SBP protein domain of SPL15. (A) Consensus DNA sequence present in the promoter region of six previously reported genes interacting with SBP DNA binding domains. CCR1-bx represents the GTAC motif binding to SBP protein CCR1 from Chlamydomonas reinhardtii[28, 29]; SBP1/2-bx1 represents the GTAC motif binding to SBP1 and SBP2 in Antirrhinum majus[30]; SPL3-bx1 and -bx2 [5, 11], SPL7-bx [32]; and SPL14-bx [31] represents the GTAC motif binding to corresponding SPL proteins from Arabidopsis thaliana. Yellow boxes, 100% conserved; green boxes, 67-83% conserved. The degree of conservation is indicated in the schematic by the height of the letters (measured as bits). (B) GTAC core repeats present in the miR156b promoter sequence (from bp -1 to -1700). (C) SDS-PAGE analysis of His-tagged recombinant SBP peptide expressed in E. coli. Lane 1, protein marker; lane 2, protein extract of non-induced E. coli cells carrying the SBP domain in pET28a; lane 3, protein extract of induced E. coli carrying the SBP domain. (D) Western blot of resolved proteins in C using anti-His antibody. (E) Electrophoretic mobility shift assay illustrating His-tagged SBP peptide bound to a labeled miR156b promoter fragment containing tandem repeats of the GTAC core. Lane 1, labeled DNA plus SBP peptide. lane 2, labeled DNA only; lane 3, labeled DNA plus SBP protein and 100-fold excess unlabeled DNA. Black arrow shows the shifted band in lane 1.
Discussion
Enhanced expression of miR156b in sk156
In this study, we highlight a new Arabidopsis mutant, sk156, with enhanced expression of miR156b due to the insertion of a T-DNA activation tag. The elevated miR156b transcript levels in sk156 are responsible for the full spectrum of morphological phenotypes observed for this mutant. Increased seed carotenoid levels and enhanced branching are consistent with phenotypes observed in B. napus expressing AtmiR156b under the control of the CaMV35S promoter [22]. These results confirm the role for miR156b in seed carotenoid accumulation, and are in agreement with morphological changes reported in earlier studies [3, 6, 12, 14, 35].
Differences in transcriptional regulation for the native miR156 promoter, the CaMV35S promoter, and the enhanced miR156b native promoter, in addition to different T-DNA insertion sites, can lead to substantial differences in transcript levels between individual transgenic lines and, consequently, to variation in phenotype. For example, miR156b transcription driven by the CaVM 35S promoter was more than 2000-fold higher than native miR156b transcription in WT plants, whereas miR156b transcription in the sk156 mutant was ~80-fold higher than in WT plants. Such differences in miR156b expression levels may result in distinct phenotypes between the sk156 mutant and 35S:miR156b lines. For example, numerous small rosettes and tiny leaves were produced in sk156 (Figure 1C), and comparatively much smaller and many more rosette leaves were noted in the 35S:miR156b lines in this and previous studies [6].
Strigolactone-independent seed carotenoid increase in sk156
Our data indicated that morphological traits and seed carotenoid profiles are affected in sk156. A link between altered carotenoid profiles and branching was previously observed in max mutants [17, 19] and in an Arabidopsis histone methyltransferase (SDG8)-defective mutant [36]. This was largely due to the effect on the biosynthesis of carotenoid-derived strigolactone branching inhibitors [20, 21]. In the current study, we also demonstrated that the elevated levels of seed carotenoids in Arabidopsis mutant sk156 were due to the enhancer-driven expression of miR156b. However, morphological phenotypes of the T1 progeny of sk156 crossed with WT (in which CCD7 and CCD8 are transcribed normally) and of transgenic WT overexpressing CCD7 or CCD8 (data not shown) remained almost the same as in sk156. Exogenous application of the artificial strigolactone GR24 (2'-epi-5-deoxystrigol) to sk156 seedlings for six weeks by supplementing the chemical to the in vitro plant growing in MS media did not affect sk156 phenotypes (data not shown). These data suggest that ectopic expression of the miR156b-induced branching phenotype in sk156 might not be directly related to the strigolactone pathway. In silico analysis revealed that no known carotenoid biosynthesis or catabolism genes in Arabidopsis possess sequences complimentary to the mature sequence of miR156b[2]. As well, transcript levels of β-ring carotenoid hydroxylase, ε-ring carotene hydroxylase, lycopene β-cyclase, lycopene ε-cyclase, phytoene desaturase, phytoene synthase or ζ-carotene desaturase were not significantly different in leaves and siliques of sk156 and WT (data not shown). Thus, miR156b likely affects the seed carotenoid pathway indirectly through the regulation of SPL networks as demonstrated in the miR156-regulated accumulation of anthocyanin via several SPL genes [10]. However, we cannot exclude the possibility of changes to seed carotenoid enzyme activities in the absence of transcription changes, such as occured with phytoene synthase in etiolated Arabidopsis[37]. Also, the impact on seed carotenoid accumulation may not necessarily be due to biosynthesis, but rather to improved carotenoid sequestration and storage as demonstrated in the Or mutant of Brassica oleracea[38]. These possibilities point to the need for additional investigation to unravel the biological basis for increased carotenoid accumulation in sk156 seeds.
In sk156, a substantial increase in seed carotenoid levels was detected in all except cryptoxanthin. This minor carotenoid, however, was reduced in sk156 seeds. Cryptoxanthin may be more efficiently converted to zeaxanthin in sk156 than in WT Arabidopsis. This possibility could arise potentially due to differences in transcription of the related genes between WT and sk156 and should be tested in the future. For example, in Zea mays, a β-carotene hydroxylase variant, ZmBCH1, converts β-carotene to β-cryptoxanthin and zeaxanthin, whereas ZmBCH2 can only convert β-carotene to β-cryptoxanthin [39]. Also, in Arabidopsis two variants of β-carotene hydroxylase were found to function differently [40].
A slight decrease in carotenoid abundance was observed in leaves of the sk156 line even though carotenoid levels were increased in sk156 seeds. Transgenic B. napus in which the gene encoding lycopene ε-cyclase was constitutively down-regulated also showed increased carotenoids in seeds, but not in leaves [41]. Different mechanisms control carotenoid metabolism in green and non-green tissues, including seeds [42–44]. In developing chloroplasts, the synthesis of carotenoids, such as lutein, β-carotene, zeaxanthin and violaxanthin, is regulated in concert with the light-regulated assembly of light-receiving antennae and photosystem centers into which the carotenoids are integrated [45, 46]. In contrast, carotenoid compounds in non-green tissue plastids vary widely in quantity and composition and their synthesis is not necessarily regulated similarly to chloroplasts. In seeds, carotenoids are stored in elaioplasts (lipid storing plastids) or amyloplasts (starch storing plastids) [42, 47]. Evaluating the impact of miR156b on the transcription of genes specifying differentiation into these carotenoid-rich plastid types could lead us to understand why miR156b regulates carotenoid accumulation differently in seeds and leaves of sk156.
SPL15 regulation of the morphology and carotenoid alterations in sk156
Our 5’ RACE assays showed that SPL15 mRNA was cleaved at three cleavage sites in the segment located near the middle of the coding sequence and is complementary to mature miR156, suggesting a role for miR156-directed cleavage in SPL15 transcript processing. However, distinct morphologies were often difficult to find between many of the Arabidopsis mutants defective in individual SPL genes both in our study and in others [8]. In the present study, spl15 mutants exhibited slightly stunted shoot growth and a modest increase in carotenoid levels, which were some (but not the full spectrum) of the phenotypes of sk156, while the other spl mutants examined in this study did not exhibit any discernible phenotypes. Consistently, phenotypes specified by the sk156 mutant were largely restored to WT phenotypes in sk156 complemented with SPL15m expressed under the control of the CaVM35S promoter or the AS1 promoter. These pieces of evidence clearly suggest that miR156 suppresses SPL15 in the sk156 mutant and causes much of its phenotypic iteration.
Other SPL genes must contribute somewhat to the sk156 mutant phenotype development, since spl15 mutants did not show the entire spectrum of sk156 phenotypes and sk156 was not fully restored to a WT phenotype by SPL15m expressed from a native SPL15 promoter. These data not only indicate that SPL15 is functionally redundant with other miR156-targeted SPL genes, but that this redundancy probably is limited by the endogenous regulation of temporal and spatial gene expression. Our data derived from AS1 promoter-driven SPL15m experiments show that the leaf primordium within the shoot apex is a crucial site for SPL15m expression to restore sk156 phenotypes. This is consistent with previous findings that the shoot apex is a predominant expression site for some SPL genes [2, 5, 13] and that AS1 promoter-driven miR156 promotes leaf initiation by suppressing SPL genes at the shoot apical meristem [14]. AS1 is expressed only in emerging lateral organ primordia, and in seeds its expression is detectable only in limited subepidermal cells corresponding to cotyledon initials at the heart stage [48]. In our hands, AS1:GUS + plants did not show visible GUS staining in seeds but did show staining in shoot tips, suggesting that SPL15m transcript level might not be significantly elevated in AS1:SPL15m+ seeds compared to sk156. The morphological changes in transgenic sk156 expressing 35S/AS1:SPL15m point to functional redundancy of SPL15 with at least SPL9 and SPL10 on plastochron length [8], SPL9 on shoot development [13], and SPL3, SPL4, and SPL5 on flowering stage transition [4, 12].
Expression of miR156b-insensitive SPL15m modifies leaf shape in the sk156 lines carrying 35S:SPL15m or SPL15p:SPL15m constructs. A role for SPL15 in leaf morphological traits was also revealed by Usami et al [49]. The enhancement of miR156b expression in sk156 possibly prevents the plants from maintaining sufficient SPL transcript levels required for normal control of leaf architecture, including those from SPL15. These findings are consistent with reduced expression of 10 SPL genes in the inflorescences of Arabidopsis lines over-expressing miR156b[6]. Expression of SPL15 gene in leaf primodia was also important for rescuing WT-like seed carotenoid and morphology traits in sk156. This was supported partially by the finding that leaf primodia are a site where SPL factors control the rate of leaf initiation [8].
SPL15 feedback regulation of miR156b
Feedback loops, in which a miRNA-targeted transcription factor either increases or decreases the expression of its cognate miRNA, have been reported in animals [50, 51]. However, the mechanism underlying this feedback regulation is unclear, particularly in plants. In our hands, complementation of sk156 with SPL15m leads to increased levels of SPL15 transcripts and decreased levels of miR156b transcripts. The reduction in miR156b expression due to increased SPL15 was consistent with the inverse correlation of SPL3 and mature miR156 transcripts in WT Arabidopsis at the early vegetative stages (19- and 26-d-old) [12]. These data suggest that negative feedback regulation by SPL15 may exist for miR156b. A recent study on the regulation of miR156a and miR172b in early vegetative stages proposed feedback regulation between the miRNAs and their targets but did not further investigate the underlying mechanism [9].
All SPL family members contain a conserved SBP DNA binding domain [5, 30] with variations outside the domain. Moreover, DNA sequences in promoters of SPL-regulated genes contain a GTAC core [5, 28, 29, 31, 32, 52], which is a key element for transcription regulation [53]. Our gel shift assay demonstrated a direct physical interaction between the SPL15 SBP domain and the promoter region of miR156b containing three repeats of the GTAC core sequence. This suggests a mechanism for the underlying feedback regulation of miR156b by its target SPL15. Support for this finding comes from reports by Birkenbihl et al [52] and Yamasaki et al [32] which indicated that such an interaction also exists between the SBP domain for SPL1, -3, -7 and -8 and the GTAC element.
Conclusions
In the present study we showcase a new mutant, sk156, in which a T-DNA insert containing four CaMV35S enhancers hyper-induced miR156b. We documented phenotypes for several sk156 lines complemented with different expression cassettes for miR156b-sensitive and miR156b-insensitive SPL15, and showed for the first time a direct interaction between SPL15 and miR156b. These data and the inverse pattern of miR156b and SPL15 expression in mutant sk156 lead us to conclude that negative feedback regulation of miR156b by SPL15 exists. A second finding is that miR156b regulates seed carotenoid abundance in Arabidopsis differently than in leaves although the underlying molecular basis remains a subject for further investigation.
Methods
Plants and growth conditions
The sk156 mutant was isolated from a T-DNA activation-tagged mutant population of Arabidopsis Col-4 [23]. The population was generated at the AAFC Saskatoon Research Centre using the pSKI015 binary vector containing a T-DNA with a Bar gene and 4 tandem CaMV enhancers [54]. SALK and FLAG T-DNA insertion lines with knockouts in SPL genes were obtained from the Arabidopsis Biological Resource Center and the FST (flanking insertion sites) project (http://www.arabidopsis.org; Additional file 2). max mutants were kindly provided by Dr. M-S Peng (University of Guelph) and transgenic Arabidopsis lines over-expressing miR156b under the control of the CaMV35S promoter were provided by Dr. R. S. Poethig (University of Pennsylvania). To recover homozygous mutant lines, segregating offspring grown were grown on half-strength Murashige-Skoog (MS) basal medium (Sigma, M5519-50L) with 0.8% agar and a selective agent (7.5 mg/L phosphinothricin or 50 mg/L kanamycin) and were screened by PCR using primers specific for each insertion site (Additional file 5). Homozygous plants were transplanted into growth chamber pots for further characterization.
For phenotypic analysis, plants were grown in 32-well flats containing Co-Co Mix soil mixture consisting of compacted coconut fibre/peat moss/vermiculite (1/3/3, v/v/v) and 15-9-12 "Osmocote PLUS" controlled release fertilizer (Scotts Company LLC). Plant age was recorded from the time seeds were imbibed. All plants were kept in a controlled environment growth chamber under a 16/8 h light/dark photoperiod with a light intensity of 230 μE/min/m2 and temperature of 20oC/17oC. Leaf and branch number means were calculated from 30 individual plants of mutant or WT lines. A Duncan's multiple range test was conducted to show significant differences between the means at p < 0.01 and p < 0.05.
Carotenoid extraction and quantification
For carotenoid analysis, leaves of 28-day-old plants were excised and samples immediately ground in liquid nitrogen. Mature seeds harvested from different plants were kept at -80oC for further analysis. Tissues were maintained at -80oC before extraction to minimize carotenoid degradation. Approximately 100 mg of leaf tissue powder and 150 mg of homogenized seeds were used for carotenoid extraction. Carotenoid extraction and HPLC quantitative analysis were carried out as previously described for seeds [41]. HPLC chromatograph peaks were identified by comparing their retention times and absorption spectra to authentic standards, and quantified using standard curves corresponding to carotenoid compounds. Pure chemical standards for β-carotene and lutein were purchased from Sigma. β-cryptoxanthin, zeaxanthin and violaxanthin were purchased from CaroteNature (Lupsingen, Switzerland). Each sample was pooled from the leaves or seeds of 6-10 different plants. Six replicates (two biological replicates with three technical replicates) were used for carotenoid analysis. A Duncan's multiple range test was used to separate statistically different means at p < 0.01 and p < 0.05.
Molecular characterization of the sk156mutant
Plant genomic DNA was extracted to determine T-DNA copy number in the sk156 mutant. Southern analysis was conducted using 15 μg of genomic DNA digested with BamHI and HindIII. The 1.4 kb fragment was excised from a pSKI015 plasmid using HindIII and EcoRI, labeled with 32P, and used as a hybridization probe. To identify the T-DNA insertion site of sk156, genomic DNA was digested with each of four blunt-end restriction enzymes provided in the GenomeWalk kit (Clontech), ligated with adaptors, and flanking regions amplified by nested PCR with the T-DNA specific primers pSKI015-GW-LB1 and pSKI015-GW-LB2 according to the manufacturer’s instructions (Additional file 5). Cloned GenomeWalk PCR products were sequenced using the pSKI015-GW-LB2 primer (Additional file 5) and the T-DNA insertion site was determined by BLAST analysis to the TAIR sequence database (Version 9) (http://www.arabidopsis.org/Blast/index.jsp). Genetic segregation of plants with T-DNA inserts was completed in triplicate by recording T-DNA insertion and the number of surviving seedlings that geminated out of ~100 seeds growing in half-strength MS media containing 7.5 mg/L phosphinotricin or 50 mg/L kanamycin, followed by χ2 analysis.
Constructs and plant transformation
Total RNA for cDNA synthesis was extracted from 3-week-old Arabidopsis thaliana ecotype Columbia plants using the QIAGEN RNeasy kit (Qiagen) according to the manufacturer’s instructions. DNase I was used for on-column DNA digestion to minimize genomic DNA contamination. First-strand cDNA was synthesized by reverse transcription of 300 ng of total RNA in a final reaction volume of 20 μL using random primers and 200 units of SuperScript™ II Reverse Transcriptase (Invitrogen). Full length cDNA of miR156b, SPL15, AT4G30975 and bHLH069 was amplified using Platinum® Taq High Fidelity DNA Polymerase (Invitrogen) and gene specific primer pairs (miR156b_XbaI_F/ miR156b_SacI_R; pXbaI_ SPL15F/ pSacI _SPL15R; p30975BamHI_F /p30975SacI_R; and pbHLH_F / pbHLH_R, respectively), with addition of restriction enzyme sites at the 5’ and 3’ ends of the genes (Additional file 5). SPL15m was generated by introducing 11 mutations into the predicted miR156 binding site using the PCR primers pSPL15m851F and pSPL15m865R containing mutated sequences (Additional file 5). The AS1 promoter was isolated from A. thaliana Columbia genomic DNA using pAS1_HindIII_F and pAS1_XbaI_R (Additional file 5) according to Wang et al [8], while a 3 kb DNA fragment upstream of the SPL15 transcription start site including both the ‘proximal promoter’ and ‘distal promoter’ regions [55] was amplified by PCR using the primers pSPL15Pr3_HindIII_F and pSPL15Pr3_XbaI_R (Additional file 5) and used as the native SPL15 promoter. Amplified PCR fragments were cloned into pCR2.1-TOPO vector (Invitrogen) and verified by DNA sequencing. The fragments were retrieved with corresponding restriction enzymes and inserted into the binary vector pBI121 digested with the same pair of restriction enzymes (replacing the original gusA gene or CaMV35S promoter in the vector). The resulting binary expression vectors contained an nptII gene and transgenes under the control of either the CaMV35S promoter, the AS1 promoter, or the native SPL15 promoter and were introduced into Agrobacterium tumefaciens GV3101pMP90 and used to transform Arabidopsis using the standard floral dip method. Putative transgenic lines that survived antibiotic selection were transplanted into the greenhouse for further analysis.
5’RACE
5’RACE was carried out using the FirstChoice RLM RACE Kit (Ambion) following the manufacturer's instructions specially for the detection of miRNA degradation products [56]. Briefly, total RNA was isolated from 3-week-old seedlings as described above. Five micrograms of total RNA was ligated to the RNA adapter and a random-primed reverse transcription reaction was performed to synthesize cDNA. A second round of PCR was carried out using a nested adapter primer and primers specific for SPL15 (Additional file 5). The RACE products were cloned into the pGEM T Easy vector (Promega) for sequencing.
Quantitative Real Time PCR analysis
Total RNA extraction, on-column genomic DNA digestion, and first-strand cDNA synthesis were conducted as described above. Real time quantitative RT-PCR (qPCR) analyses were performed with gene specific primers listed in supplemental Table S1. To quantify miR156b, primers based on the stem sequence of the pre-miR156b hairpin structure were designed to measure premature miR156b according to Schmittgen et al. [57] (Additional file 5). qPCR mixtures contained 10 μl of diluted cDNA, 12.5 μl of 2X SYBR Green qPCR Master Mix (Cat No. 11735-040, Invitrogen) and 200 nM of each gene-specific primer in a final volume of 25 μl. qPCR reactions were conducted using the StepOne Plus system and software (Applied Biosystems) using a relative standard curve method and default reaction parameters. The relative index of the standard curve was over 98%. Control PCR reactions with no templates were also performed for each primer pair. The specificity of amplicons was verified by melting curve analysis (60 to 95oC) after 40 cycles and by agarose gel electrophoresis. All samples were assayed in triplicate from two independent RNA preparations. Mean expression values of all replicates were calculated and normalized to the expression of PEROXIN 4 (PEX4, set at 1), a suitable endogenous reference gene because of its stable and low level expression in Arabidopsis[58]. All PCR reactions displayed efficiencies between 87 and 115%. Normalized means were analyzed for significant differences by a Duncan’s test (p < 0.05).
Expression and extraction of SBP-domains
For recombinant protein expression in Escherichia coli, a coding sequence fragment encoding the SPL15 SBP domain (80 amino acid residues) plus 5 amino acid residues both upstream and downstream [48] was amplified by PCR using the primers p15SBP-BamHI-F and p15SBP-SalI-R with addition of BamHI at the 5’ end and SalI and a TGA stop codon at the 3’ end (Additional file 5). The PCR product was digested with BamHI and SalI and cloned into the pET28a vector (Novagen) between the BamHI and SalI sites. The resulting chimeric recombinant protein fused with a His tag (16.3kD) was expressed in E. coli strain Rosetta 2(DE3) pLacI (Clontech) by induction with 0.8 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested by centrifugation and resuspended in BugBuster® Master Mix (Novagen) to lyse the cells. Recombinant His-tagged SBP proteins were detected in the inclusion bodies using anti-His antibody (G-18) (Santa Cruz Biotechnology) in Western blot assays. Proteins in inclusion bodies were solubilized with 1.5% sarkosyl (N-laurylsarcosine) according to Frangioni and Neel [59], and recombinant SBP protein recovered and re-folded using a protein refolding kit (TB234, Novagen) according to the manufacturer’s instructions. Protein concentration was measured in lysates by a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) and proteins separated in 13% SDS–polyacrylamide gels. The protein was stored at 4°C until use in gel shift assays.
Electrophoretic mobility shift assays
The ability of SPL15 SBP to bind to the miR156b promoter DNA was examined using electrophoretic mobility shift assays. A 46 residue DNA fragment, which included the three repeated GTAC core sequences close to the transcriptional start site of miR156b (shown in Figure 8B), was synthesized using primers R156b-bx1-U and R156b-bx1-L (Additional file 5), hybridized and labeled with digoxigenin using a DIG Gel Shift kit (second generation) (Roche). The labeled DNA fragment (~20 fmol) was incubated for 30 min at 25°C with or without the SPL15 SBP protein (~500 ng) in 20 μL of reaction buffer containing 10 mM Tris (pH 7.5), 50 mM KCl, 5 mM MgCl, 5 mM DTT, 2.5% glycerol, and 0.05% NP-40. Then, a 100-fold excess of the unlabeled promoter DNA fragment was added to the reactions. After incubation, the mixtures were separated by polyacrylamide electrophoresis (7.5% gel) at 4oC for 1.5 h (0.8 V/cm2) in 0.5 X TBE (44.5 mM Tris base, 44.5 mM boric acid, and 1 mM EDTA at pH 8.0). DNA was blotted onto a nylon membrane and mobility changes for the labeled miR156b promoter fragment detected with digoxigenin-specific antibodies.
Authors’ information
Submitting author: Shu Wei, PhD in Plant Sciences and currently Professor in Plant Molecular Biology and Biotechnology at Anhui Agricultural University, China, working on, but not limited to plant microRNA regulated gene networks and metabolic pathway engineering.
References
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA: Conservation and divergence of plant microRNA genes. Plant J. 2006, 46: 243-259. 10.1111/j.1365-313X.2006.02697.x.
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP: Prediction of plant microRNA targets. Cell. 2002, 110: 513-520. 10.1016/S0092-8674(02)00863-2.
Xie K, Wu C, Xiong L: Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142: 280-293. 10.1104/pp.106.084475.
Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P: The miRNA156/157 recognition element in the 3’UTR of the Arabidopsis SBP box geneSPL3prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49: 683-693. 10.1111/j.1365-313X.2006.02983.x.
Cardon G, Höhmann S, Klein J, Nettesheim K, Sardler H, Huijser P: Molecular characterisation of theArabidopsisSBP-box genes. Gene. 1999, 237: 91-104. 10.1016/S0378-1119(99)00308-X.
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D: Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005, 8: 517-527. 10.1016/j.devcel.2005.01.018.
Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J: Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008, 418: 1-8. 10.1016/j.gene.2008.03.016.
Wang JW, Czech B, Weigel D: miR156-Regulated SPL transcription factors define an endogenous flowering pathway inArabidopsis thaliana. Cell. 2009, 138: 738-749. 10.1016/j.cell.2009.06.014.
Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS: The sequential action of miR156 and miR172 regulates developmental timing inArabidopsis. Cell. 2009, 138: 750-759. 10.1016/j.cell.2009.06.031.
Gou J-Y, Felippes FF, Liu C-J, Weigel D, Wang J-W: Negative regulation of anthocyanin biosynthesis inArabidopsisby a miR156-targeted SPL transcription factor. Plant Cell. 2011, 23: 1512-1522. 10.1105/tpc.111.084525.
Cardon G, Höhmann S, Nettesheim K, Saedler H, Huijser P: Functional analysis of theArabidopsis thalianaSBP-box geneSPL3: A novel gene involved in the floral transition. Plant J. 1997, 12: 367-377. 10.1046/j.1365-313X.1997.12020367.x.
Wu G, Poethig RS: Temporal regulation of shoot development inArabidopsis thalianabymiR156and its targetSPL3. Development. 2006, 133: 3539-3547. 10.1242/dev.02521.
Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P: The microRNA regulated SBP-box genesSPL9andSPL15control shoot maturation in Arabidopsis. Plant Mol Biol. 2008, 67: 183-195. 10.1007/s11103-008-9310-z.
Wang JW, Schwab R, Czech B, Mica E, Weigel D: Dual effects of miR156-targetedSPLgenes andCYP78A5/KLUHon plastochron length and organ size inArabidopsis thaliana. Plant Cell. 2008, 20: 1231-1243. 10.1105/tpc.108.058180.
Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M: Arabidopsis SBP-box genesSPL10,SPL11andSPL2control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009, 50: 2133-2145. 10.1093/pcp/pcp148.
Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY: Temporal control of trichome distribution by microRNA156-targetedSPLgenes inArabidopsis thaliana. Plant Cell. 2010, 22: 2322-2335. 10.1105/tpc.109.072579.
Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O: MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol. 2004, 14: 1232-1238. 10.1016/j.cub.2004.06.061.
Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg S, Turnbull C, Srinivasan P, Goddard P, Leyser O: MAX1encodes a cytochrome P450 family member that acts downstream ofMAX3/4to produce a carotenoid-derived branch-inhibiting hormone. Del Cell. 2005, 8: 443-449. 10.1016/j.devcel.2005.01.009.
Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, Leyser O: MAX4andRMS1are orthologous dioxygenase-like genes that regulate shoot branching inArabidopsisand pea. Genes Dev. 2003, 17: 1469-1474. 10.1101/gad.256603.
Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S: Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008, 455: 195-200. 10.1038/nature07272.
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H, Bécard G, Beveridge CA, Rameau C, Rochange SF: Strigolactone inhibition of shoot branching. Nature. 2008, 455: 189-194. 10.1038/nature07271.
Wei S, Yu B, Gruber MY, Khachatourians GG, Hegedus DD, Hannoufa A: Enhanced seed carotenoid levels and branching in transgenicBrassica napusexpressing theArabidopsis miR156bgene. J Agric Food Chem. 2010, 58: 9572-9578. 10.1021/jf102635f.
Robinson SJ, Tang LH, Mooney BA, McKay SJ, Clarke WE, Links MG, Karcz S, Regan S, Wu YY, Gruber MY, Cui D, Yu M, Parkin IA: An archived activation tagged population ofArabidopsis thalianato facilitate forward genetics approaches. BMC Plant Biol. 2009, 9: 101-10.1186/1471-2229-9-101.
Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ: Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45: 982-993. 10.1111/j.1365-313X.2006.02666.x.
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC: The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003, 20: 735-747. 10.1093/molbev/msg088.
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G: Arabidopsistranscription factors: genome-wide comparative analysis among eukaryotes. Science. 2000, 290: 2105-2110. 10.1126/science.290.5499.2105.
Eshed Y, Baum SF, Perea JV, Bowman JL: Establishment of polarity in lateral organs of plants. Curr Biol. 2001, 11: 1251-1260. 10.1016/S0960-9822(01)00392-X.
Quinn JM, Barraco P, Eriksson M, Merchant S: Coordinate copper- and oxygen-responsiveCyc6andCpx1expression inChlamydomonasis mediated by the same element. J Biol Chem. 2000, 275: 6080-6089. 10.1074/jbc.275.9.6080.
Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S: A regulator of nutritional copper signaling inChlamydomonasis an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci U S A. 2005, 102: 18730-18735. 10.1073/pnas.0507693102.
Klein J, Saedler H, Huijser P: A new family of DNA binding proteins includes putative transcriptional regulators of theAntirrhinum majusfloral meristem identity geneSQUAMOSA. Mol Gen Genet. 1996, 250: 7-16.
Liang X, Nazarenus TJ, Stone JM: Identification of a consensus DNA-binding site for theArabidopsis thalianaSBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry. 2008, 47: 3645-3653. 10.1021/bi701431y.
Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T: SQUAMOSApromoter binding protein-like7 is a central regulator for copper homeostasis inArabidopsis. Plant Cell. 2009, 21: 347-361. 10.1105/tpc.108.060137.
Crooks G, Hon G, Chandonia JM, Brenner SE: WebLogo: a sequence logo generator. Genome Res. 2004, 14: 1188-1190. 10.1101/gr.849004.
Schwab R: Functions and target selection of Arabidopsis microRNAs. 2006, Tübingen Germany: Eberhard Karls University, PhD thesis
Chuck G, Cigan AM, Saeteurn K, Hake S: The heterochronic maize mutantCorngrass1results from overexpression of a tandem microRNA. Nat Genet. 2007, 39: 544-549. 10.1038/ng2001.
Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ: Regulation of carotenoid composition and shoot branching inArabidopsisby a chromatin modifying histone methyltransferase, SDG8. Plant Cell. 2009, 21: 39-53. 10.1105/tpc.108.063131.
Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M: Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009, 60: 424-435. 10.1111/j.1365-313X.2009.03966.x.
Paolillo DJ, Garvin DF, Parthasarathy MV: The chromoplasts of Or mutants of cauliflower (Brassica oleracea. Var. Botrytis). Protoplasma. 2004, 224: 245-253. 10.1007/s00709-004-0059-1.
Li Q, Farre G, Naqvi S, Breitenbach J, Sanahuja G, Bai C, Sandmann G, Capell T, Christou P, Zhu C: Cloning and functional characterization of the maize carotenoid isomerase and β-carotene hydroxylase genes and their regulation during endosperm maturation. Transgenic Res. 2010, 19: 1053-1068. 10.1007/s11248-010-9381-x.
Tian L, DellaPenna D: Characterization of a second carotenoid βhydroxylase gene from Arabidopsis and its relationship to theLUT1locus. Plant Mol Biol. 2001, 47: 379-388. 10.1023/A:1011623907959.
Yu B, Lydiate DJ, Schäfer UA, Hannoufa A: Characterization of a β-carotene hydroxylase ofAdonis aestivalisand its expression inArabidopsis thaliana. Planta. 2007, 226: 181-192. 10.1007/s00425-006-0455-1.
Howitt CA, Pogson BJ: Carotenoid accumulation and function in seed and non-green tissues. Plant Cell Environ. 2006, 29: 435-445. 10.1111/j.1365-3040.2005.01492.x.
Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J: A chromoplast-specific carotenoid biosynthesis revealed by cloning of the tomatowhite-flowerlocus. Plant Cell. 2006, 18: 1947-1960. 10.1105/tpc.105.039966.
Lopez AB, Van Eck J, Conlin BJ, Paolillo DJ, O'Neill J, Li L: Effect of the cauliflowerOrtransgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. J Exp Bot. 2008, 59: 213-223. 10.1093/jxb/erm299.
Young AJ: Factors that affect the carotenoid composition of higher plants and algae. Carotenoids in photosynthesis. Edited by: Young AJ, Britton G. London: Chapman and Hall, 1993:161-205.
Römer S, Lübeck J, Kauder F, Steiger S, Adomat C, Sandmann G: Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab Eng. 2002, 4: 263-272. 10.1006/mben.2002.0234.
Vishnevetsky M, Ovadis M, Vainstein A: Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci. 1999, 4: 232-235. 10.1016/S1360-1385(99)01414-4.
Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA: Asymmetric leaves1mediates leaf patterning and stem cell function inArabidopsis. Nature. 2000, 408: 967-971. 10.1038/35050091.
Usami T, Horiguchi G, Yano S, Tsukaya H: Themore and smaller cellsmutants ofArabidopsis thalianaidentify novel roles forSQUAMOSA PROMOTER BINDING PROTEIN-LIKEgenes in the control of heteroblasty. Development. 2009, 136: 955-964. 10.1242/dev.028613.
Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A: A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007, 317: 1220-1224. 10.1126/science.1140481.
Varghese J, Cohen SM: microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling inDrosophila. Genes Dev. 2007, 21: 2277-2282. 10.1101/gad.439807.
Birkenbihl RP, Jach G, Saedler H, Huijser P: Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 2005, 352: 585-596. 10.1016/j.jmb.2005.07.013.
Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg M, Hippler M, Merchant S: Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell. 2002, 14: 673-688. 10.1105/tpc.010420.
Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J: Activation tagging in Arabidopsis. Plant Physiol. 2000, 122: 1003-1014. 10.1104/pp.122.4.1003.
Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV: PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 2003, 31: 114-117. 10.1093/nar/gkg041.
Wang XJ, Reyes JL, Chua NH, Gaasterland T: Prediction and identification ofArabidopsis thalianamicroRNAs and their mRNA targets. Genome Biol. 2004, 5: R65-10.1186/gb-2004-5-9-r65.
Schmittgen TD, Jiang J, Liu Q, Yang L: A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004, 32: e43-10.1093/nar/gnh040.
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR: Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139: 5-17. 10.1104/pp.105.063743.
Frangioni JV, Neel BG: (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993, 210: 179-187. 10.1006/abio.1993.1170.
Acknowledgements
We thank Dr. Mingsheng Peng, University of Guelph, for kindly providing us with the max mutants and Dr. R. S. Poethig, University of Pennsylvania, for transgenic Arabidopsis plants over-expressing the miR156b gene. GR24 (2'-epi-5-deoxystrigol) was a kind gift from Professor S. Yamaguchi at the RIKEN Plant Science Center, Japan and Prof. M. Sasaki at Kumamoto University, Japan. This work was supported by the Genome Canada-funded project “Designing Oilseeds for Tomorrow’s Markets”.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors' contributions
SW designed the experiments and conducted the majority of the experimentation, analyzed data, and drafted the manuscript; IAP Parkin constructed and provided the Arabidopsis activation-tagged population; BY was involved in construction of SPL15 binary vectors; MJG performed computational sequence analysis of the miR156b promoter binding motif; MYG, and AH provided critical feedback on experimental concepts. MYG, GGK, DDH and AH revised the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12870_2012_1160_MOESM1_ESM.png
Additional file 1: Schematic diagram of disrupted carotenoid cleavage dioxygenase genes CCD7 and CCD8 in the max3-9 and max4-1 mutants used in this study. Boxes represent exons and lines represent introns. Triangles show the T-DNA insertion sites. The max mutants were previously reported by (Booker et al. 2004). (PNG 54 KB)
12870_2012_1160_MOESM3_ESM.png
Additional file 3: Reduced lengths of cauline stem basal internodes in three spl15 mutants compared with WT Arabidopsis . Length of the cauline stem basal inter-node was measured from the rosette core up to the first visible basal node for WT, three spl mutants and sk156 plants grown for 6 weeks. (PNG 34 KB)
12870_2012_1160_MOESM4_ESM.png
Additional file 4: Confirmation by PCR of transgene presence in sk156 lines transformed with a 35S:SPL15m gene. The miR156 insensitive SPL15m contained 11 mutated nucleotides as described in Materials and Methods. Primer sequences are listed in Additional file 5. (A) Activation-tag from pSKI015 T-DNA detected in transgenic sk156 plants carrying 35S:SPL15m cassette (lanes 0-11) and in the sk156 background alone (lanes 12 and 13). Primers SK2222-F (430bp upstream) and SK2222-R (1830bp downstream) flanking the T-DNA insertion site and primer pSKI015-GW-LB2 (439bp to the T-DNA left border) were used to detect the insert. In WT lane, no T-DNA insert was detected and only a fragment close to 1.9kb was present due to genomic DNA amplified with the primers flanking the T-DNA insertion site. In homozygous sk156 plants which did not carry 35S:SPL15m, a single T-DNA fragment (869bp) was generated. 1kb, 1-kb Plus DNA ladder (Invitrogen); WT, Col-4; pSKI015, plasmid containing the activation tag present in sk156. (B) Transgene SPL15m detected in the transgenic sk156 lines carrying 35S:SPL15m (lanes 0-14), but not in sk156 alone using primers 35SF3 and SPL15R. (PNG 543 KB)
12870_2012_1160_MOESM6_ESM.png
Additional file 6: PCR confirmation of transgene presence in four different miR156-sensitive or miR156-insensitive transgenic Arabidopsis populations used in this manuscript. (A) Lanes 2-8, PCR product (818bp) for 7 transgenic plants carrying a 35S:AT4G30975 cassette in a WT background using primers 35S-F3 and p795-3R. P, binary plasmid pBI121 containing 35S:AT4G30975 as a positive control. WT, Col-4. (B), (C) and (D) Transgene PCR product (636bp) carrying AS1:SPL15m, AS1:SPL15n and SPL:SPL15m cassettes in a sk156 background, respectively, using primers SPL15-871F and NosTer-R6. P, plasmid containing 35S:SPL15 as a positive control. Black arrows points to the DNA marker. Primer sequences are listed in Additional file 5. 1kb, 1-kb Plus DNA ladder (Invitrogen). (PNG 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wei, S., Gruber, M.Y., Yu, B. et al. Arabidopsis mutant sk156 reveals complex regulation of SPL15 in a miR156-controlled gene network. BMC Plant Biol 12, 169 (2012). https://doi.org/10.1186/1471-2229-12-169
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-12-169