Abstract
Background
The cultivated potato (Solanum tuberosum L.) is an important food crop, but highly susceptible to many pathogens. The major threat to potato production is the Irish famine pathogen Phytophthora infestans, which causes the devastating late blight disease. Potato breeding makes use of germplasm from wild relatives (wild germplasm) to introduce resistances into cultivated potato. The Solanum section Petota comprises tuber-bearing species that are potential donors of new disease resistance genes. The aim of this study was to explore Solanum section Petota for resistance genes and generate a widely accessible resource that is useful for studying and implementing disease resistance in potato.
Description
The SolRgene database contains data on resistance to P. infestans and presence of R genes and R gene homologues in Solanum section Petota. We have explored Solanum section Petota for resistance to late blight in high throughput disease tests under various laboratory conditions and in field trials. From resistant wild germplasm, segregating populations were generated and assessed for the presence of resistance genes. All these data have been entered into the SolRgene database. To facilitate genetic and resistance gene evolution studies, phylogenetic data of the entire SolRgene collection are included, as well as a tool for generating phylogenetic trees of selected groups of germplasm. Data from resistance gene allele-mining studies are incorporated, which enables detection of R gene homologs in related germplasm. Using these resources, various resistance genes have been detected and some of these have been cloned, whereas others are in the cloning pipeline. All this information is stored in the online SolRgene database, which allows users to query resistance data, sequences, passport data of the accessions, and phylogenic classifications.
Conclusion
Solanum section Petota forms the basis of the SolRgene database, which contains a collection of resistance data of an unprecedented size and precision. Complemented with R gene sequence data and phylogenetic tools, SolRgene can be considered the primary resource for information on R genes from potato and wild tuber-bearing relatives.
Similar content being viewed by others
Background
Potato ranks third on the list of economically important food crops world-wide. However, potato is susceptible to many diseases and as a consequence, potato production depends on the application of enormous amounts of pesticides. The major disease in potato is late blight, which is caused by the oomycete pathogen Phytophthora infestans [1]. A durable control strategy based on natural resistance to late blight is of great importance.
Fortunately, ample genetic resistance is present in wild tuber-bearing Solanum species that belong to section Petota. The section Petota contains wild species that are distributed from the southwestern USA to central Argentina and adjacent Chile [2, 3]. Potato breeders make use of this resource to introgress desired traits into cultivars [4–6]. Thus far, resistance (R) genes conferring resistance to P. infestans (Rpi) have been isolated from only a few wild Solanum species, i.e. S. demissum, S. bulbocastanum and S. venturii [7–13], and most of the resources in Solanum section Petota remain unexploited. While potato resistance breeding has so far been relatively unsuccessful, new approaches are emerging that use knowledge of effectors that are recognized by R proteins [14]. Late blight resistance as well as defeat of previously introgressed R genes is now better understood, and knowledge of effectors is being utilized in breeding and R gene deployment [15]. For consequent effector-based modern approaches, multiple R genes are required.
We have explored Solanum section Petota for R genes to P. infestans. Seeds from Solanum accessions were sown and individual genotypes are clonally maintained. This is in contrast to genebanks that maintain accessions as seeds. The rationale for our genotype-based studies is that many accessions are genetically highly dissimilar, since the majority of Petota species are self-incompatible out-breeders and heterozygous for many traits [2, 16]. We tested the Solanum genotypes for resistance to a diverse set of well-characterized P. infestans strains. Indeed we found that in many cases variation of resistance occurs within accessions, and that resistant as well as susceptible genotypes occur, e.g. in S. acaule accession 425. Quantitative resistance data from routine disease tests using three different inoculation methods [17–19] were collected and stored in a database. Also pictures of the phenotypes observed in late blight field trials were included. This resulted in a unique data set of unprecedented size and precision.
For scientific as well as breeding purposes, it is important to have good insight in the taxonomy of relevant germplasm. However, in the Solanum section Petota, various taxonomic problems occur [2, 3, 20]. To resolve the phylogenetic relationships in the SolRgene collection, all genotypes were subjected to a phylogenetic analysis based on AFLP [20, 21]. A searchable interactive NJ tree is included in SolRgene and permits identifying related germplasm for genetic studies and analyzing R genes evolution in comparison with species evolution.
R genes can be isolated using various strategies. Map-based cloning is a classic and thorough method to isolate R genes in potato which has proven successful for various Rpi genes, such as R1, R2 and R3a [7, 8, 22–24]. Allele-mining is a more high-throughput strategy to isolate genetic variants of R gene homologues, among which functional R genes can be detected. Strongly supported with rapid growing sequence information on R genes in the potato and tomato [25–32], efficient allele-mining for R gene homologues (RGHs) is dependent on availability of phenotyped genetic material, such as the SolRgene collection. Recently, effector genomics is emerging as an efficient tool to accelerate R gene cloning, often in combination with small-scale genetic mapping and allele-mining [14].
Genetic studies are facilitated by generating populations. By making sexual crosses between resistant and susceptible Solanum genotypes, experimental (segregating) populations were produced. These are the basis for genetics studies that can lead to map-based cloning. For example, S. venturii was crossed with S. neorossii and the generated segregating population (7663) was used for the map-based cloning of Rpi-vnt1.3 [12]. Such cloned natural R genes are indicated as cisgenes if they originate from the potato plant itself or from crossable species. Due to the highly heterozygous and cross-pollinating nature of potato, genetic modification would be a major step in quickly achieving resistance by either using transgenesis or cisgenesis approaches. Cisgenesis is the combination of marker free transformation with only cisgenes [33].
The SolRgene database was developed to provide a comprehensive dataset that can be used to explore R genes to potato pathogens in the Solanum section Petota. Major effort was attributed to Rpi genes acting against the late blight disease, but also other R genes were studied. A vast collection of disease phenotyping data, genetic data, allele-mining data of resistance genes against potato pathogens, and phylogenetic data complemented with an interactive tree tool are included, and are useful to unravel the genetic variation of R genes in the Petota gene pool. Hence, SolRgene can be considered the primary resource for information on R genes of Solanum for the scientific community and potato breeders.
Construction and content
Data source of accessions
The current database version contains information on 1061 accessions (Table 1), obtained from different genebanks, i.e. the The Dutch-German Potato Collection at the Centre for Genetic Resources The Netherlands (CGN), The Commonwealth Potato Collection (CPC), The Groß Lüsewitz Potato Collection (GLKS), The potato Collection of the Vavilov Institute (VIR), The Potato Collection of the International Potato Center (CIP), and The US Potato Genebank (NRS). Accessions were originally collected from 14 countries in South and Central America, i.e. Argentina, Bolivia, Brazil, Chile, Colombia, Costa Rica, Ecuador, Guatemala, Mexico, Paraguay, Peru, USA, Uruguay and Venezuela, and geographical collection data are all included.
The accessions represent Solanum section Petota and a few outgroup species. From these accessions, a set of 5009 genotypes was obtained from seeds, which are clonally maintained in vitro and are available upon request. The prevalence of 15 R genes or QTL is analyzed in the plant collection, with links to published papers.
Phylogeny
Previously, we constructed a neighbor-joining (NJ) tree for 4929 genotypes [21]. Related to that dataset, we offer an interactive, searchable version of this NJ tree in SolRgene. The different groups in the tree can be highlighted using the three letter species codes [34]. Neighbor-joining tree's, for a selected subsets of genotypes, can be calculated on-the-fly. The complete SolRgene germplasm collection is also classified according to Hawkes [2], except for a few interspecific hybrids that were generated across series (Table 1).
Crossability in tuber-bearing Solanumspecies
A total of 1032 successful crosses were made and information is stored within the SolRgene database. The crosses were produced within and/or between species. In most cases, crosses within designated phylogenetic species groups were successful.
Genotype-based resistance information
From each accession, on average five genotypes were characterized and the data of 5009 wild Solanum genotypes were stored independently in the database. Resistance data were generated using three different inoculation methods, i.e. high-throughput in vitro assays [17], detached leaf assays [18] and multi-year field trials [19] (Table 1). Pages explaining the disease assessment protocols supported with photographs are included. The majority (3936 genotypes) of the wild Solanum collection was tested with P. infestans isolate 90128 in the high throughput inoculation assay on in vitro plants. Part of the genotypes was tested in the laboratory using a routine detached leaf assay (1367 genotypes) with the P. infestans isolates 90128, IPO-C, or both. Solanum genotypes were also tested in field trials (986 genotypes) in 2005, 2007, or both, with P. infestans isolate IPO-C. A graphical representation of the resistance data facilitates a quick overview. From the field trails, 694 photographic images displaying symptoms on 362 genotypes are presented. Also two time lapse pictures of disease progress in the field are shown. Altogether, phenotyping of resistance resulted in 5 major sets of quantitative resistance data on the wild Solanum genotypes, and averages are presented too.
In addition to genotypes originating from genebank accessions, 7602 offspring genotypes from the generated populations were assessed for P. infestans resistance (see below). The offspring genotypes were generally tested in detached leaf tests, with four well-characterized P. infestans isolates, i.e. 90128, IPO-C, 88069, and H30P04, the reference isolate of the P. infestans genome sequence [35]. These resistance data provide information on segregation of specific Rpi genes.
Map-based cloning using SolRgene
To generate the required segregating populations for genetic mapping, resistant and susceptible plants were crossed. In total 1032 populations were generated. From these, 188 populations were phenotyped for resistance, and data were included in SolRgene. Populations that are suitable for map-based cloning show a clear segregation between resistance and susceptibility in the F1, the so-called black & white segregation. Several of such segregating populations have entered into a pipe-line of genetic mapping and R gene isolation in our laboratory [12, 14, 32, 36–38]. The first R genes to P. infestans (Rpi) from this resource have recently been cloned, such as Rpi-vnt1.1, Rpi-vnt1.3 [12] and Rpi-sto1 [14].
Allele-mining in SolRgene
Mining for late blight resistance genes Rpi-vnt1, Rpi-blb1, Rpi-blb2, Rpi-blb3 and its homolog R2 on the SolRgene collection led to identification of an extensive number of RGH. Some of these were found functional and confer resistance to P. infestans [23, 37–40]. A similar strategy was employed to identify four novel functional Rx genes (Rx3, Rx4, Rx5 and Rx6) from distinct Solanum species [41], which confer extreme resistance to PVX and share high sequence homology with Rx1 and Rx2.
Database and web application
SolRgene has been designed for simple and efficient data retrieval. It is composed of two major components: a relational database created using MySQL 5.1 and a web application which is implemented using PHP 5.2.6. The web interface runs on the Apache 2 Web server and is hosted on a Debian lenny linux server. The PHP scripts dynamically execute complex SQL queries to retrieve data from the database according to user criteria. HTML output, formatted using CSS style sheets, is generated to display results to the end-users. The relational MySQL database schema is segmented into seven main entities: Accession information, genotype information, population information, experiment information, disease observations, R genes and allele mining. Supporting tables were implemented containing e.g. information on origin of the accessions and availability of a genotype in vitro. Photos of many of the accessions are stored. Hyperlinks to Google Scholar and the NCBI gene bank records are provided for obtaining additional information on each accession/genotype. The Google Earth API is used to show the original collection site of the accession/group of selected accessions. All stored data is publicly accessible, so no authentication mechanism is built into the website.
Utility and discussion
Database web interface
SolRgene provides an interactive web-based graphical user-friendly interface to explore Solanum section Petota genotypes for resistance to P. infestans and late blight R genes. On every page, a menu tool bar appears, from which the germplasm can be searched for availability and overview data, resistance data, allele-mining data, and phylogeny. A menu for background information is also included (About). The genotype-based datasets of SolRgene provide accurate data and allow direct phenotype - genotype associations that can be made from the various menu's (Figure 1).
Schematic representation of the data mining flow of the Sol R gene database. The genotype is the central entity which links to all other types of data stored within the SolRgene database (boxes with solid lines). Links to external resources are also provided (dashed boxes). The arrows between the different boxes show the directions in which the resource can be mined. The SolRgene database can be searched using each entity as a starting point, and the resource can be mined consecutively in an iterative manner.
The germplasm can directly be searched for genotypes, accessions, species, or phylogenetic classifications via the germplasm menu. Accessions can be searched, by passport data from different genebanks, or from visual geographic locations in Google Map (Figure 1). Outputs include lists with available germplasm and whether resistance data, populations are present, and whether R genes or RGH were amplified. An integrated hyperlink to Google Scholar enables quick searches for additional information on selected accessions on the world-wide web. Populations can be searched from the diverse available Solanum species in the germplasm menu. A phylogenetic tree, can be calculated on-the-fly, from selected genotypes.
The majority of the SolRgene collection was phenotyped for resistance to P. infestans, and direct searches for resistance data can be performed via the Phytophthora resistance menu.
How to get to Rgenes?
After identifying resistant germplasm, genetics approaches are required to test whether the observed resistance can be attributed to R genes. For map-based cloning approaches, segregating populations can be selected and subjected to large-scale recombinant screenings. As an example, we present the cloning of Rpi-vnt1.3 [4] (Figure 2). First germplasm is screened for resistance to P. infestans isolates, and selected resistant genotypes are crossed with susceptible genotypes. These can be chosen using the phylogenetic tool. Obtained populations are tested for segregation of resistance to P. infestans isolates. Populations that clearly segregate for a specific R gene are used for map-based cloning purposes, sometimes in combination with allele-mining [4]. Allele-mining data for various R genes acting against P. infestans (Rpi), i.e. R2, Rpi-vnt1, Rpi-blb1, are included in SolRgene and can be linked to relevant phenotyping data. In addition, allele-mining data for resistance genes against other R genes are included, i.e. Rx that confers resistance to potato virus X (PVX). Users can take advantage of all this information, and easily link their own plant material to SolRgene, since R genes often originate from geographically restricted areas (Figure 3). Related germplasm may be identified using the Google Earth and passport data of genebanks. Also, related R genes are often identified in phylogenetically related material [40], and for this feature, the implemented phylogenetic tool can be applied.
Representation of cloning of Rpi-vnt1.3 using Sol R gene. A) Resistant Solanum germplasm is selected based on screenings with P. infestans isolates. Results to isolate IPO-C are presented. Graphic representation facilitates quick overview of the quantitative resistance data: the red indicator shows the resistance level that ranges from fully susceptible (0, left) to fully resistant (9, right). B) Resistant genotypes (365_1) are crossed with related susceptible genotypes (735_2), selected from the phylogenetic tree. C) Population 7663 (365_1 × 735_2) is segregating for resistance to P. infestans isolate IPO-C, which is visualized by the frequency distribution for the offspring. The progeny part that contains Rpi-vnt1.3 is highly resistant (right bar, resistance level 8), whereas the progeny that lacks Rpi-vnt1.3 is moderately susceptible (left bar, resistance level 3). D) After genetic mapping, the Rpi-vnt1.3 was cloned, and used for allele-mining. In this menu, R genes and related sequences can be retrieved.
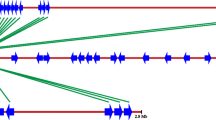
Geographic origin of Sol R gene collection and identified late blight R genes. A) The complete SolRgene collection is originating from 14 countries in South, Central and North America. B) So-far identified R genes that confer resistance to P. infestans originate mainly from mountain regions in Central and South America. C) Accessions that contain Rpi-vnt1 homologs are restricted to Argentina.
Future developments
The number of identified R genes to P. infestans and various other potato pathogens is increasing. In the near future, R genes and R gene allele-mining sequences will continue to be added (a.o. [32]), and thus, SolRgene will provide an ever increasing source of Solanum-broad sequence information. Also, we welcome sequences or other contributions from the community to be added in this database. SolRgene will also be linked to various databases including the full potato genome sequence in which our laboratory plays a leading role [25, 27, 42]. In addition, since the DNA sequence homology across Solanum species is high and ancestral R gene sequences are shared, also data for any other Solanum crops species like tomato, pepper and eggplant can be accommodated in the near future.
Conclusions
So far, no survey involving such a large number of genotypes as well as phylogenetic coverage of tuber-bearing Solanum has been made assessable, uncovering numerous new resistance sources. SolRgene is the first database that extends from phenotypic characterization to genetic dissection of the resistance by identification of functional R genes, and is regarded as the basis for potato R genes in the future. SolRgene is easily searchable through a website interface and valuable for the scientific community as well as for applied breeding. The accurate genotypic data and the continued progress towards genetic analysis and R gene isolation distinguishes SolRgene from gene bank databases. Essentially SolRgene bridges the gap between well characterized plant material oriented databases and molecular sequence databases. In the near future, R genes, R gene allele-mining sequences, and AFLPs will continue to be added. Thus, the database will provide an ever increasing source of Solanum-broad sequence information, and linked to various databases including the full potato genome sequence.
Availability and requirements
The SolRgene database is freely accessible at http://www.plantbreeding.wur.nl/SolRgenes.
Abbreviations
- R :
-
resistance
- Rpi :
-
resistance to P. infestans
- AFLP:
-
Amplified Fragment Length Polymorphism.
References
Fry W: Phytophthora infestans: the plant (and R gene) destroyer. Molecular Plant Pathology. 2008, 9 (3): 385-402. 10.1111/j.1364-3703.2007.00465.x.
Hawkes JG: The potato: evolution, biodiversity, and genetic resources.London: Belhaven Press; 1990.
Spooner DM, van den Berg RG, Rodriguez A, Bamberg J, Hijmans RJ, Lara Cabrera SI: Systematic Botany Monographs. Wild potatoes (Solanum section Petota; Solanaceae)of North and Central America. Ann Arbor, Michigan: The American Society of Plant Taxonomists; 200468
van Berloo R, Hutten RCB: Peditree: pedigree database analysis and visualization for breeding and science. Journal of Heredity. 2005, 96 (4): 465-468. 10.1093/jhered/esi059.
van Berloo R, Hutten RCB, van Eck HJ, Visser RGF: An online potato pedigree database resource. Potato Research. 2007, 50: 45-57. 10.1007/s11540-007-9028-3.
The European Cultivated Potato Database. [http://www.europotato.org]
Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C: The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant Journal. 2002, 30 (3): 361-371. 10.1046/j.1365-313X.2001.01292.x.
Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Zhang N, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF: Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. The Plant Journal. 2005, 42: 251-261. 10.1111/j.1365-313X.2005.02365.x.
van der Vossen E, Sikkema A, te Lintel Hekkert B, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S: An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant Journal. 2003, 36 (6): 867-882. 10.1046/j.1365-313X.2003.01934.x.
van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S: The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant Journal. 2005, 44 (2): 208-222. 10.1111/j.1365-313X.2005.02527.x.
Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PRJ, Visser RGF, Jacobsen E, et al: Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Molecular Plant-Microbe Interactions. 2009, 22 (6): 630-641. 10.1094/MPMI-22-6-0630.
Pel MA, Foster SJ, Park TH, Rietman H, van Arkel G, Jones JD, Van Eck HJ, Jacobsen E, Visser RGF, Van Der Vossen EAG: Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Molecular Plant-Microbe Interactions. 2009, 22 (5): 601-615. 10.1094/MPMI-22-5-0601.
Foster SJ, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, van der Vossen EAG, Jones JDG: Rpi-vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. Molecular Plant-Microbe Interactions. 2009, 22 (5): 589-600. 10.1094/MPMI-22-5-0589.
Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh S-K, Wang M, Bouwmeester K, Vosman B, Visser RGF, et al: Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE. 2008, 3 (8): e2875-10.1371/journal.pone.0002875.
Vleeshouwers VGAA, Raffaele S, Vossen J, Champouret N, Oliva R, Segretin ME, Rietman H, Cano LM, Lokossou AA, Kessel GJT, et al: Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol. 2011, 49.
Ross H: Potato breeding - Problems and perspectives. Berlin, Hamburg:Verlag Paul Parey; 1986.
Huang S, Vleeshouwers VGAA, Visser RGF, Jacobsen E: An accurate in vitro assay for high-throughput disease testing to Phytophthora infestans in potato. Plant Disease. 2005, 89: 1263-1267. 10.1094/PD-89-1263.
Vleeshouwers VGAA, van Dooijeweert W, Keizer LCP, Sijpkes L, Govers F, Colon LT: A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. European Journal of Plant Pathology. 1999, 105 (3): 241-250. 10.1023/A:1008710700363.
Colon LT, Budding DJ: Resistance to late blight (Phytophthora infestans) in ten wild Solanum species. Euphytica 1988, Supplement: 77-86.
Jacobs MMJ, Smulders MJM, van den Berg RG, Vosman B: What's in a name; Genetic structure in Solanum section Petota studied using population-genetic tools. BMC Evolutionary Biology. 2011, 11 (42).
Jacobs MMJ, van den Berg RG, Vleeshouwers VGAA, Visser M, Mank R, Sengers M, Hoekstra R, Vosman B: AFLP analysis reveals a lack of phylogenetic structure within Solanum section Petota. BMC Evolutionary Biology. 2008, 8 (1): 145-10.1186/1471-2148-8-145.
Gebhardt C, Valkonen JP: Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol. 2001, 39: 79-102. 10.1146/annurev.phyto.39.1.79.
Lokossou AA, Rietman H, Wang M, Krenek P, van der Schoot H, Henken B, Hoekstra R, Vleeshouwers VGAA, van der Vossen EAG, Visser RGF, et al: Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. Molecular Plant-Microbe Interactions. 2010, 23 (9): 1206-1216. 10.1094/MPMI-23-9-1206.
Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, et al: High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992, 132 (4): 1141-1160.
Visser RGF, Bachem CWB, de Boer JM, Bryan GJ, Chakrabati SK, Feingold S, Gromadka R, van Ham RCHJ, Huang S, Jacobs JME, et al: Sequencing the potato genome: Outline and first results to come from the elucidation of the sequence of the world's third most important food crop. American Journal of Potato Research. 2009, 86: 417-429. 10.1007/s12230-009-9097-8.
van Os H, Andrzejewski S, Bakker E, Barrena I, Bryan GJ, Caromel B, Ghareeb B, Isidore E, de Jong W, van Koert P, et al: Construction of a 10,000-marker ultradense genetic recombination map of potato: Providing a framework for accelerated gene isolation and a genomewide physical map. Genetics. 2006, 173 (2): 1075-1087. 10.1534/genetics.106.055871.
Potato Genome Sequencing Consortium. [http://www.potatogenome.net/index.php/Main_Page]
Ultra dense recombination map of potato. [http://www.plantbreeding.wur.nl/potatomap/]
Center for Biosystems Genomics. [http://www.cbsg.nl]
Meyer S, Nagel A, Gebhardt C: PoMaMo--a comprehensive database for potato genome data. Nucl Acids Res. 2005, 33 (suppl_1): D666-670.
Tomato genome sequence. [http://solgenomics.wur.nl/genomes/Solanum_lycopersicum/genome_data.pl]
Bakker E, Borm TJA, Prins P, van der Vossen EAG, Uenk G, Arens M, De Boer JM, Van Eck HJ, Muskens M, Vossen J, et al: A genome-wide genetic map of NB-LRR disease resistance loci in potato. Theor Appl Genet.
Schouten HJ, Krens FA, Jacobsen E: Cisgenic plants are similar to traditionally bred plants: international regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep. 2006, 7 (8): 750-753. 10.1038/sj.embor.7400769.
Simmonds NW: Abbreviations of potato names. European Potato Journal. 1962, 6 (3): 186-190.
Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al: Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009, 461 (7262): 393-398. 10.1038/nature08358.
Verzaux E: Resistance and susceptibility to late blight in Solanum: gene mapping, cloning and stacking. PhD thesis Wageningen University; 2010.
Champouret N: Functional genomics of Phytophthora infestans effectors and Solanum resistance genes. PhD thesis Wageningen University; 2010.
Jacobs MMJ, Vosman B, Vleeshouwers VGAA, Visser RG, Henken B, van den Berg RG: A novel approach to locate Phytophthora infestans resistance genes on the potato genetic map. Theoretical and Applied Genetics. 2010, 120 (4): 785-796. 10.1007/s00122-009-1199-7.
Wang M, Allefs S, van den Berg RG, Vleeshouwers VGAA, van der Vossen EAG, Vosman B: Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theoretical and Applied Genetics. 2008, 116 (7): 933-943. 10.1007/s00122-008-0725-3.
Pel MA: Mapping, isolation and characterization of genes responsible for late blight resistance in potato. PhD thesis Wageningen University; 2010.
Butterbach P: Molecular evolution of the disease resistance gene Rx in Solanum. PhD thesis Wageningen University; 2007.
Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J, et al: Genome sequence and analysis of the tuber crop potato. Nature. 2011, 475 (7355): 189-195. 10.1038/nature10158.
Acknowledgements and Funding
We acknowledge the Centre for Biosystems Genomics (CBSG), the Dutch Ministry of Agriculture, Nature and Food Quality (LNV427 Paraplu-plan Phytophthora) and Wageningen UR Plant Breeding for financing. We thank Guus Heselmans, Paul Heeres, Marielle Muskens, Sjefke Allefs, Robert Graveland, and Jeroen van Soestbergen for their advices and contributions to generating this resource, Patrick Butterbach, Anoma Lokossou and Miqia Wang for contributing to allele mining and Francine Govers and Geert Kessel for providing P. infestans isolates.
The acronym AFLP is a registered trademark (AFLP©) of Keygene N.V. and the AFLP© technology is covered by patents and patent applications of Keygene N.V.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
VGAAV designed the project, designed a relational database, integrated the data, and wrote the manuscript; RF worked on development and implementation of the web database, web layouts and contributed to writing the manuscript; DB carried out the majority of the technical work of disease testing in laboratory and field; MV carried out the vitro culturing Solanum plants and disease testing in vitro; MMJJ contributed to phylogenetic analysis based on AFLP; RvB for implementing the phylogenetic tools; MP, NC, PK and EB carried out the allele-mining of Rpi-vnt1, R2, Rpi-blb1 and Rx, respectively; HR contributed to field trials and generated the field photographs; DJH contributed to generating segregating populations; RH contributed Solanum accessions and information; AG contributed to Rx mining and writing the manuscript; BV contributed to phylogenetic analysis and writing the manuscript; EJ contributed to potato introgression breeding and provided valued discussions; RGFV conceived of the study, participated in its design and helped draft the manuscript. All authors have read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Vleeshouwers, V.G., Finkers, R., Budding, D. et al. SolRgene: an online database to explore disease resistance genes in tuber-bearing Solanumspecies. BMC Plant Biol 11, 116 (2011). https://doi.org/10.1186/1471-2229-11-116
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-11-116