Abstract
Background
Flatfish metamorphosis involves major physiological and morphological changes. Due to its importance in aquaculture and as a model for developmental studies, some gene expression studies have focused on the understanding of this process using quantitative real-time PCR (qRT-PCR) technique. Therefore, adequate reference genes for accurate normalization are required.
Results
The stability of 12 potential reference genes was examined during larval development in Senegalese sole (Solea senegalensis) and Atlantic halibut (Hippoglossus hippoglossus) to determine the most suitable genes for qRT-PCR analysis. Transcription levels of genes encoding β-Actin (ACTB), glyceraldehyde-3P-dehydrogenase (GAPDH), annexin A2 (ANXA2), glutathione S-transferase (GST), ornithine decarboxylase (ODC), hypoxanthine phosphoribosyltransferase (HPRT1), ubiquitin (UBQ), elongation factor 1 alpha (eEF1A1), 18S ribosomal RNA, and the ribosomal proteins S4 (RPS4) and L13a (RPL13a) were quantitated. Two paralogous genes for ACTB were analyzed in each of both flatfish species. In addition, two paralogous genes for GAPDH were studied in Senegalese sole. RPL13a represented non-orthologous genes between both flatfish species. GeNorm and NormFinder analyses for expression stability revealed RPS4, UBQ and eEF1A1 as the most stable genes in Senegalese sole, Atlantic halibut and in a combined analysis. In all cases, paralogous genes exhibited differences in expression stability.
Conclusion
This work suggests RPS4, UBQ, and eEF1A1 genes as useful reference genes for accurate normalization in qRT-PCR studies in Senegalese sole and Atlantic halibut larvae. The congruent results between both species in spite of the drastic differences in larval development suggest that selected housekeeping genes (HKGs) could be useful in other flatfish species. However, the finding of paralogous gene copies differentially expressed during development in some HKGs underscores the necessity to identify orthologous genes.
Similar content being viewed by others
Background
Pleuronectiformes (flatfish) are a broad taxonomic group comprising 11 families and about 500 species worldwide, and some of them are of high commercial interest both in fisheries and aquaculture. Senegalese sole, Solea senegalensis, and Atlantic halibut, Hippoglossus hippoglossus, are two representative species of Pleuronectidae and Soleidae families, respectively. Pleuronectidae (right-eye flounders) and Soleidae (soles) families include 93 species in 39 genera and 89 species in 22 genera, respectively [1, 2]. All these species share in common an asymmetrical body development and a bottom-dwelling mode of life. Although they possess bilateral body symmetry during the larval stages, they undergo drastic morphological and physiological changes in order to become an asymmetric benthic juvenile. Senegalese sole and Atlantic halibut show clear differences in the timing of larval development and metamorphosis (Figure 1). In Atlantic halibut, hatching occurs approximately two weeks after fertilization. The yolk-sack period extends to 46–50 days after hatching (DAH) and the metamorphosis begins very late with the migration of the left eye about 80 DAH, taking 2–3 weeks [3]. In contrast, Senegalese sole hatches 3 days after fertilization and first feeding occurs at 3 DAH. Metamorphosis takes only one week between 12 and 19 DAH [4]. In spite of the differences in the timing of metamorphosis, this process is mediated by thyroid hormones (THs) in both species [5, 6]. Because of the intrinsically interesting role in development as well as the subsequent effects on characteristics such as growth, deformities or malpigmentations, which are important in aquaculture, flatfish metamorphosis has been the subject of gene expression studies using PCR-based methods [7–9]. However, adequate and reliable reference genes for accurate quantification remain to be validated.
Quantitative real-time PCR has become one of the most widespread techniques for mRNA gene expression analysis due to its accuracy, broad dynamic range, sensitivity and reproducibility [10–12]. Although absolute and relative quantitation approaches are possible, the latter is preferred due to the normalization of sample variations using internal housekeeping genes (HKGs). These HKGs act as endogenous controls that allow for the correction of experimental variations caused by pipetting errors, inhibitory compounds, reverse transcription (RT) efficiency or quality of starting material [11]. Traditionally, highly expressed genes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB) or 18S ribosomal RNA have been considered as putative HKGs. Due to their key role in metabolism, cytoskeleton and ribosome structure, respectively, it is generally assumed that they are expressed at a constant level in different tissues, cells or experimental treatments. However, there is increasing evidence that their expression can vary during development or in response to external treatments [13–15]. In addition, some of them (i.e. GAPDH and other glycolytic enzymes as well as ribosomal proteins) possess functional paralogous genes that exhibit tissue-specific expression patterns (muscle, liver and/or brain) [16–19]. As an alternative, the use of multiple internal control genes have been proposed [20]. Different algorithms have been developed to choose the most appropriate HKG combination set [20–22]. However, candidate reference genes need to be validated under the specific experimental conditions being evaluated.
The development of large-scale genomics of Senegalese sole and Atlantic halibut (Pleurogene project; [23]) has made possible the availability of a large number of EST sequences. We have identified commonly used reference genes in both species. Twelve candidate HKGs for gene expression analysis during larval developmental in Senegalese sole and Atlantic halibut have been investigated. The possible use of the selected HKGs for gene expression studies in other flatfish species is discussed.
Results
mRNA transcription levels of potential HKGs during larval development
A total of 12 genes were evaluated as potential HKGs in Senegalese sole and Atlantic halibut. Owing to the widespread use of GAPDH and ACTB as reference genes, different paralogous genes identified in Senegalese sole and Atlantic halibut were included in the analysis. Only ACTB1 and GAPDH1 were considered orthologs in Senegalese sole and Atlantic halibut as determined by phylogenetic analysis and gene expression profile in tissues. In addition, RPL13a genes were paralogous copies between the two flatfish species. ODC could not be detected in Senegalese sole EST libraries and thus it was only analyzed in Atlantic halibut.
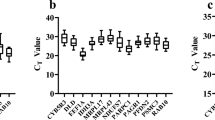
The Ct value indicates the fractional cycle at which the fluorescence intensity equals the threshold fluorescence. Ct values are inversely related with the abundance of a specific transcript in the sample and they have to be converted to a linear form by using the amplification efficiency to estimate gene expression changes. Ct values of HKGs in Senegalese sole ranged between 11.05 (18S rRNA) and 27.20 (RPL13a) (Figure 2). In Atlantic halibut, Ct values varied between 7.35 (18S rRNA) and 30.16 (ANXA2). HPRT1, ANXA2 and GST were expressed at low levels in both flatfish species (median Ct values above 20 cycles). The smallest Ct variation was exhibited by GAPDH2 (1.25) and 18S rRNA (1.80) in the Senegalese sole and by 18S rRNA (1.25), ACTB1 (2.45) and HPRT1 (2.61) in the halibut. On the contrary, RPL13a (4.40), GAPDH1 (3.45) and ACTB1 (3.45) showed the most variable expression levels in the sole, whereas ANXA2 (7.34) and GAPDH1 (5.99) were the most variable in the halibut.
The mRNA expression profiles for all HKGs studied in Senegalese sole and Atlantic halibut during larval development are depicted in Figures 3 and 4. ANXA2 exhibited the most pronounced variation in Atlantic halibut since expression levels dropped 6.75 Cts from hatching to 57 DAH. In Senegalese sole, RPL13a showed the highest Ct variation (3.95) between 3 and 13 DAH.
Gene expression stability analysis
Expression stability for each HKG during larval development in Senegalese sole and Atlantic halibut was analyzed using the geNorm and NormFinder software. GeNorm (or pairwise comparison approach) ranks HKGs according to the similarity of the expression profiles across the sample set. All investigated genes exhibited high average expression stability (M) values (lower than 1 in both species). In the Senegalese sole, ACTB2, RPS4, GAPDH2, UBQ and eEF1A1 were the most stable genes with M values lower than 0.5. In Atlantic halibut, HPRT1, ACTB2, UBQ, eEF1a1, RPS4 and RPL13a exhibited M values lower than 0.5 (Figure 5). Additionally, a combined analysis to determine the most stable genes in both flatfish was carried out. In this analysis, only the putative orthologous genes in both flatfish species were included. Although higher M values were observed, ACTB1, RPS4, UBQ and eEF1A1 showed M values lower than 0.6.
To determine the minimum number of HKGs necessary for an accurate normalization, a pairwise variation Vn/n+1 analysis was performed (Figure 6). If we take into account the default cut-off value of 0.15 [24], geometric averaging of 3 genes in Senegalese sole and 2 in Atlantic halibut would be required for accurate normalization. Combined analysis yielded a V2/3 value of 0.138 although the inclusion of a third reference gene reduced slightly the pairwise variation (V3/4 = 0.132).
NormFinder (Model-based approach) ranks the best candidate HKGs according to their minimal combined inter- and intragroup expression variation [21]. Results of NormFinder analysis for our data are shown in Table 1. In Senegalese sole, GAPDH2, RPS4, ACTB2, eEF1A1 and UBQ appeared as the most stable genes (stability between 0.219 and 0.289). In Atlantic halibut, RPS4, UBQ, RPL13a, GST, HPRT1 and eEF1A1 showed the highest stability values (0.159–0.348). These HKG data sets agree with those obtained using geNorm with slight differences in the ranking order. Combined analysis showed RPS4, UBQ and eEF1A1 as the most stable genes (0.278–0.413)
Discussion
Flatfish metamorphosis is characterized by drastic morphological and physiological changes that determine the switch from a pelagic to a benthic lifestyle. During this period, erythrocyte populations and myosin light chains change to adult types [25, 26], gastric glands differentiate [27] and an asymmetrical pigmentation develops [8, 28]. Owing to the interest of larval stages from flatfish in developmental studies and their impact on essential characteristics of interest in aquaculture (digestion, pigmentation, growth, etc), some gene expression surveys have focused on this developmental period. However, appropriate HKGs that ensure reliable and accurate mRNA quantitation are required. To our knowledge, this is the first study that evaluates the suitability of potential HKGs for gene expression analysis in flatfish larvae. Twelve potential HKGs in the pleuronectid Atlantic halibut and the soleid Senegalese sole were investigated. The high similarity for the most stable HKGs in spite of the differences in the timing of metamorphosis (12–19 DAH in Senegalese sole, and 80–100 DAH in Atlantic halibut) and metamorphic stages (S1–S4 in Senegalese sole [4], and stages 6–9 [29] in Atlantic halibut), gives support and highlight the suitability for gene expression studies in other flatfish.
Two different analytical approaches were used to study the most suitable HKGs during larval development. The pairwise comparison approach (geNorm) selects the most suitable HKGs on the basis of the variation of expression ratios between HKGs expression across sample set. Genes with the lowest M (internal control gene-stability measure) values have the most stable expression [20]. However, the geNorm algorithm is highly dependent on the assumption that none of the genes being analyzed are co-regulated. Although most HKGs analyzed in this study are involved in different cellular functions, such co-regulation cannot be ruled out since some genes of the translational apparatus have been analyzed and the broad effect of THs on metabolism and flatfish metamorphosis [5, 30, 31]. Hence, HKG expression data were also analyzed using a model-based approach (NormFinder). This methodology ranks candidate genes on the basis of the minimal inter- and intragroup variation [21]. Both experimental approaches identified a similar HKG subset as the most stable. Although only two and three genes were necessary for accurate normalization in Senegalese sole and Atlantic halibut, respectively, and only two in the combined analysis as determined by GeNorm, the inclusion of one additional gene clearly improved the pairwise variation in all cases. Hence, RPS4, UBQ and eEF1A1, involved in protein biosynthesis and degradation, appeared as the most suitable HKG set for accurate normalization in gene expression studies during larval development in flatfish.
Three rounds of large-scale gene duplications have been identified in fish [19, 32]. These duplications are responsible, at least in part, for the speciation, the adaptive radiations and the high morphological complexity of fish [33]. The majority of these gene duplicates are lost or silenced during evolution, and only in some instances they can either acquire new functions or divide the ancestral original functions [19, 34]. Several paralogous genes including pseudogenes have been described for glycolytic enzymes [18, 19], ribosomal proteins [16, 35], or elongation factor 1 alpha [14], some of which are routinely considered as potential HKGs. Our data demonstrate that expression profiles and stability vary between paralogous gene copies. GAPDH2 exhibited higher gene stability than GAPDH1 in Senegalese sole. Similarly, ACTB2 showed higher stability than ACTB1 in both species as determined both with geNorm and NormFinder approaches. If we compare the ranking of the two species, the RPL13a paralogous genes were ranked differentially in the two species. In contrast, ACTB1 and GAPDH1 orthologous genes were ranked in a similar way. These data underscore the necessity for a phylogenetic analysis to demonstrate orthology in order to select the appropriate HKG in other flatfish and to make expression profiles comparable between species. In addition, the design of primers is a key point. Due to the high similarity in the coding sequence between some paralogous genes, it becomes necessary to confirm that the paralog of interest is amplified by sequencing of the PCR products or by designing the primers in divergent regions of the UTRs. The simultaneously cross-amplification of different paralogous genes that are differentially expressed would render a consensus expression profile that would make the comparisons among species difficult.
GAPDH has been traditionally considered as a HKG due to its key role in the glycolytic pathway. However, GAPDH is associated with several functions during evolution such as membrane fusion, microtubule bundling, phosphotransferase activity, nuclear RNA export, DNA replication and repair, apoptosis, age-related neurodegenerative disease, prostate cancer and viral pathogenesis (reviewed in [36]). There is increasing evidence that the expression of this gene is modified during development, in tissues and by physiological conditions [13–15]. Our expression data ranked GAPDH1 (muscle-specific isoform) as an inadequate HKG in both Senegalese sole and Atlantic halibut. These data agree with recent findings that have demonstrated a down-regulation of GAPDH1, contrary to GAPDH2, by THs [18], a key regulator of metamorphosis in the two species [5, 6]. This differential regulation between both GAPDH paralogous genes would explain the higher expression stability of GAPDH2 (brain-specific isoform), ranked as the best HKG by NormFinder, and the inadequacy of GAPDH1 as HKG during larval development in the two flatfishes.
Conclusion
Our results indicate that RPS4, eEF1A1 and UBQ genes may be good candidate reference genes for larval development studies in flatfish as selected by geNorm and NormFinder. However, the application of these genes to achieve an accurate normalization in gene expression studies in other species requires the identification of putative orthologous genes.
Methods
Source of fish and experimental rearing conditions
All experimental Senegalese sole larvae were obtained from fertilized eggs collected from breeding tanks where breeders spawned naturally under ambient conditions. Eggs were incubated at a density of 2000 eggs L-1 in 300 L cylinder conical tanks with gentle aeration and one water exchange every two hours. Temperature and salinity during all experiments were 20°C and 38 ppt, respectively. Newly hatched larvae were transferred to a 400 L tank at an initial density from 45 to 50 larvae L-1 with a 16L:8D photoperiod and a light intensity of 600–800 lux. Larvae were fed rotifers (Brachionus plicatilis) between 3 DAH and 9 DAH. From 7 DAH enriched brine shrimp (Artemia sp.) metanauplii were fed until the end of the experiment. Pools of larvae from 2 to 22 DAH (n = 3) were collected, washed with DEPC water, frozen in liquid nitrogen and stored at -80°C until analysis.
Atlantic halibut larval samples were obtained from Scotian Halibut Limited (Clark's Harbour, Nova Scotia, Canada). Fertilized eggs and hatched larvae were incubated at 6 – 7°C until 5 – 6 weeks of age after hatching. After this stage, larvae were raised at 10.0 – 11.2°C. The samples were taken at 1 DAH (hatching), 33, 48, 57, 68, 82, 92, and 103. Sampled larvae were anaesthetized in overdosed tricaine methane sulfonate (TMS, Syndel Laboratories, Vancouver, BC, Canada), and then, transferred to RNAlater (Ambion, Austin, TX, USA) and kept overnight at 4°C. The samples were removed from RNAlater and individually stored in a 1.6 ml microfuge tube at -80°C until RNA was isolated. Ten, 5, and 2 individuals were pooled to isolate total RNA from samples at 1, 33, and 48 DAH, respectively. One whole fish was used to obtain total RNA at other stages. Three RNA samples were obtained for each stage.
Identification of HKG cDNAs in Senegalese sole and Atlantic halibut
The potential HKGs evaluated in this study were: β-Actin (ACTB), glyceraldehyde-3P-dehydrogenase (GAPDH), annexin A2 (ANXA2), glutathione S-transferase (GST), ornithine decarboxylase (ODC), hypoxanthine phosphoribosyltransferase (HPRT1), ubiquitin (UBQ), elongation factor 1 alpha (eEF1A1), 18S ribosomal RNA (18S rRNA), and the ribosomal proteins S4 (RPS4) and L13a (RPL13a). Expressed sequence tags (ESTs) encoding HKGs from Senegalese sole and Atlantic halibut were identified after sequence analysis of normalized cDNA libraries constructed from different larval stages and adult tissues. ODC was only identified in H. hippoglossus. Two different genes encoding for GAPDH (referred to as GAPDH1 and GAPDH2) [18] and for ACTB (referred to as ACTB1 and ACTB2) were identified in both species. Similarly, two RPL13a genes were identified in Senegalese sole. RPL13a-like1 was orthologous of RPL13a gene in halibut. However, to compare gene expression stability between paralogs, the RPL13a-like2 (referred to as RPL13a in the text), probably a pseudogene, was analyzed in this study. Sequences for each HKG in both flatfish species were aligned to assess orthology. GAPDH1 and ACTB1 sequences were orthologs in Senegalese sole and Atlantic halibut. In contrast, ACTB2 and RPL13a were paralogs between both flatfish species. GAPDH2 only was studied in Senegalese sole. Accession numbers and main function of each HKG evaluated are indicated in Table 2.
RNA isolation and gene expression analysis
Homogenization of Senegalese sole larvae was carried out using Lysing Matrix D (Q-BioGene Inc., Carlsbad, CA, USA) for 40 s at speed setting 6 in the Fastprep FG120 instrument (Bio101, Thermo Savant Instruments). Total RNA was isolated from 50 mg of pooled larvae using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). For Atlantic halibut larvae, RNeasy Fibrous Tissue Mini Kit (Qiagen) was used to isolate total RNA. Fish were transferred into a 1.5 ml microfuge tube with RLT buffer from the kit and disrupted with a pellet pestle (Kimble-Kontes, Vineland, NJ). The mixture was passed through 18- and 21-gauge needles, to homogenize the sample and to reduce the viscosity. All RNA isolation procedures were performed in accordance with the manufacturer's protocol. In all cases, total RNA was treated twice with DNase I using the RNase-Free DNase kit (Qiagen) for 30 min for Senegalese sole or DNA-free (Ambion, Austin, TX, USA) for Atlantic halibut in order to avoid amplification of genomic DNA. For Senegalese sole RNA, sample quality was checked using Experion (Bio-Rad) and quantification was performed spectrophotometrically. Total RNA (1 μg) from each sample was reverse-transcribed using the iScript™ cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA). Lack of genomic DNA contamination was confirmed by PCR amplification of RNA samples in the absence of cDNA synthesis.
Real-time analysis was carried out on an iCycler (Bio-Rad). Reactions were performed in a 25 μL volume containing cDNA generated from 10 ng of original RNA template, 300 nM each of specific forward (F) and reverse (R) primers (Table 3), and 12.5 μl of iQ™ SYBR Green Supermix (Bio-Rad). Matching oligonucleotide primers were designed using Oligo v 6.89 software (Medprobe, Oslo, Norway) or primer 3 (Whitehead Institute for Biomedical Research). The amplification protocol used was as follows: initial 7 min denaturation and enzyme activation at 95°C, 40 cycles of 95°C for 15 s and 70°C for 30 s, and for Atlantic halibut, initial 7 min denaturation and enzyme activation at 95°C, 40 cycles of 95°C for 20 s, 58°C for 20 s and 72°C for 20 s. Each assay was performed in duplicate. To estimate efficiencies, a standard curve was generated for each primer pair based on known quantities of cDNA (10-fold serial dilutions corresponding to cDNA transcribed from 100 to 0.01 ng of total RNA). All calibration curves exhibited correlation coefficients higher than 0.99 and the corresponding real-time PCR efficiencies were in the range 0.86–1.01 (Table 3).
In order to ensure comparability among data, the threshold values among plates were normalized by manually setting the Ct values corresponding to at least two previous reference samples. Raw Ct values were transformed to quantities using the comparative Ct method and specific efficiencies [37]. Data obtained were converted into correct input files, according to the requirements of the software, and analyzed using geNorm [20], and NormFinder [21].
References
Froese R, Pauly D: FishBase. World Wide Web electronic publication 2007. version (01/2007) [http://www.fishbase.org]
Helfman G, Collette B, Facey D: The diversity of fishes. Blackwell Science 1997.
Einarsdottir IE, Silva N, Power DM, Smaradottir H, Bjornsson BT: Thyroid and pituitary gland development from hatching through metamorphosis of a teleost flatfish, the Atlantic halibut. Anat Embryol 2006, 211: 47-60. 10.1007/s00429-005-0055-z
Fernández-Díaz C, Yúfera M, Cañavate JP, Moyano FJ, Alarcón FJ, Díaz M: Growth and physiological changes during metamorphosis of Senegal sole reared in the laboratory. J Fish Biol 2001, 58: 1-13. 10.1111/j.1095-8649.2001.tb00557.x
Manchado M, Infante C, Asensio E, Planas JV, Cañavate JP: Thyroid hormones down-regulate thyrotropin β subunit and thyroglobulin during metamorphosis in the flatfish Senegalese sole (Solea senegalensis Kaup). Gen Comp Endocrinol 2008, 155: 447-455. 10.1016/j.ygcen.2007.07.011
Solbakken JS, Norberg B, Watanabe K, Pittman K: Thyroxine as a mediator of metamorphosis of Atlantic halibut ( Hippoglossus hippoglossus L.). Environ Biol Fish 1999, 56: 53-65. 10.1023/A:1007542526040
Bai J, Solberg C, Fernandes JM, Johnston IA: Profiling of maternal and developmental-stage specific mRNA transcripts in Atlantic halibut Hippoglossus hippoglossus . Gene 2006, 386: 202-210. 10.1016/j.gene.2006.09.012
Campinho MA, Silva N, Sweeney GE, Power DM: Molecular, cellular and histological changes in skin from a larval to an adult phenotype during bony fish metamorphosis. Cell Tissue Res 2007, 327: 267-284. 10.1007/s00441-006-0262-9
Funes V, Asensio E, Ponce M, Infante C, Cañavate JP, Manchado M: Insulin-like growth factors I and II in the sole Solea senegalensis : cDNA cloning and quantitation of gene expression in tissues and during larval development. Gen Comp Endocrinol 2006, 149: 166-172.
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, et al.: The real-time polymerase chain reaction. Mol Aspects Med 2006, 27: 95-125. 10.1016/j.mam.2005.12.007
Bustin SA, Benes V, Nolan T, Pfaffl MW: Quantitative real-time RT-PCR-a perspective. J Mol Endocrinol 2005, 34: 597-601. 10.1677/jme.1.01755
Bustin SA, Nolan T: Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech 2004, 15: 155-166.
Aoki T, Naka H, Katagiri T, Hirono I: Cloning and characterization of glyceraldehyde-3-phosphate dehydrogenase cDNA of Japanese flounder Paralichthys olivaceus . Fisheries Sci 2000, 66: 737-742. 10.1046/j.1444-2906.2000.00120.x
Olsvik PA, Lie KK, Jordal AEO, Nilsen TO, Hordvik I: Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol Biol 2005, 6: 21. 10.1186/1471-2199-6-21
Zhang Z, Hu J: Development and validation of endogenous reference genes for expression profiling of medaka ( Oryzias latipes ) exposed to endocrine disrupting chemicals by quantitative real-time RT-PCR. Toxicol Sci 2007, 95(2):3563-3568.
Karsi A, Patterson A, Feng J, Liu Z: Translational machinery of channel catfish: I. A transcriptomic approach to the analysis of 32 40S ribosomal protein genes and their expression. Gene 2002, 291: 177-186. 10.1016/S0378-1119(02)00595-4
Manchado M, Infante C, Asensio E, Cañavate JP, Douglas SE: Comparative sequence analysis of the complete set of 40S ribosomal proteins in the Senegalese sole (Solea senegalensis Kaup) and Atlantic halibut ( Hippoglossus hippoglossus L.) (Teleostei: Pleuronectiformes): phylogeny and tissue- and development-specific expression. BMC Evol Biol 2007, 7: 107. 10.1186/1471-2148-7-107
Manchado M, Infante C, Asensio E, Cañavate JP: Differential gene expression and dependence on thyroid hormones of two glyceraldehyde-3-phosphate dehydrogenases in the flatfish Senegalese sole ( Solea senegalensis Kaup). Gene 2007, 400: 1-8. 10.1016/j.gene.2007.05.008
Steinke D, Hoegg S, Brinkmann H, Meyer A: Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol 2006, 4: 16. 10.1186/1741-7007-4-16
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002, 3: RESEARCH0034. 10.1186/gb-2002-3-7-research0034
Andersen CL, Jensen JL, Orntoft TF: Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004, 64: 5245-5250. 10.1158/0008-5472.CAN-04-0496
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP: Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 2004, 26: 509-515. 10.1023/B:BILE.0000019559.84305.47
Pleurogene project[http://pleurogene.ca/]
Vaerman JL, Saussoy P, Ingargiola I: Evaluation of real-time PCR data. J Biol Regul Homeost Agents 2004, 18: 212-214.
Miwa S, Inui Y: Thyroid hormone stimulates the shift of erythrocyte populations during metamorphosis of the flounder. J Exp Biol 1991, 259: 222-228.
Yamano K, Takano-Ohmuro H, Obinata T, Inui Y: Effect of thyroid hormone on developmental transition of myosin light chains during flounder metamorphosis. Gen Comp Endocrinol 1994, 93: 321-326. 10.1006/gcen.1994.1036
Miwa S, Yamano K, Inui Y: Thyroid hormone stimulates gastric development in flounder larvae during metamorphosis. J Exp Biol 1991, 261: 424-430.
Tagawa M, Aritaki M: Production of symmetrical flatfish by controlling the timing of thyroid hormone treatment in spotted halibut Verasper variegatus . Gen Comp Endocrinol 2005, 141: 184-189. 10.1016/j.ygcen.2005.01.013
Sæle Ø, Solbakken JS, Watanabe K, Hamre K, Power D, Pittman K: Staging of Atlantic halibut ( Hippoglossus hippoglossus L.) from first feeding through metamorphosis, including cranial ossification independent of eye migration. Aquaculture 239: 445-465.
Miwa S, Tagawa M, Inui Y, Hirano T: Thyroxine surge in metamorphosing flounder larvae. Gen Comp Endocrinol 1988, 70: 158-163. 10.1016/0016-6480(88)90105-0
Schreiber AM, Specker JL: Metamorphosis in the summer flounder ( Paralichthys dentatus ): stage-specific developmental response to altered thyroid status. Gen Comp Endocrinol 1998, 111: 156-166. 10.1006/gcen.1998.7095
Volff JN: Genome evolution and biodiversity in teleost fish. Heredity 2005, 94: 280-294. 10.1038/sj.hdy.6800635
Blomme T, Vandepoele K, de Bodt S, Simillion C, Maere S, Van de Peer Y: The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol 2006, 7(5):R43. 10.1186/gb-2006-7-5-r43
Lynch M, Conery JS: The evolutionary fate and consequences of duplicate genes. Science 2000, 290: 1151-1155. 10.1126/science.290.5494.1151
Patterson A, Karsi A, Feng J, Liu Z: Translational machinery of channel catfish: II. Complementary DNA and expression of the complete set of 47 60S ribosomal proteins. Gene 2003, 305: 151-160. 10.1016/S0378-1119(02)01183-6
Sirover MA: New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta 1999, 1432: 159-184.
Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCtMethod. Methods 2001, 25: 402-408. 10.1006/meth.2001.1262
Acknowledgements
This work is part of the "Pleurogene" project funded as one of Genome Canada-Genoma España collaborative projects. We thank Scotian Halibut Limited, especially Ms. Melissa Rommens and Ms. Shelley LeBlanc for kindly providing halibut samples. MM is the recipient of a research contract from INIA cofunded by European Social Fund.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors' contributions
CI carried out the gene expression analysis in Senegalese sole and helped to draft the manuscript MPM carried out the gene expression analysis in halibut, carried out sequence analyses and helped to draft the manuscript. EA performed the Senegalese sole cultures and samplings. JPC participated in the study design and helped to draft the manuscript. MR participated in the study design and drafted the manuscript. MM designed the study, carried out sequence analyses, and drafted the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Infante, C., Matsuoka, M.P., Asensio, E. et al. Selection of housekeeping genes for gene expression studies in larvae from flatfish using real-time PCR. BMC Molecular Biol 9, 28 (2008). https://doi.org/10.1186/1471-2199-9-28
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2199-9-28