Abstract
Background
We determined the prevalence and evidence for physical linkage amongst integrons, insertion sequences, Tn21 and Tn7 transposons in a collection of 1327 E. coli obtained over a 19-year period from patients in Kenya.
Results
The prevalence of class 1 integrons was 35%, class 2 integrons were detected in 3 isolates but no isolate contained a class 3 integron. Integron lacking the 3’-CS or those linked to sul3 gene or IS26 or those containing the ISCR1 were only detected in multidrug resistant (MDR) strains. The dfrAs were the most common cassettes and their prevalence was: - dfrA1(28%), dfrA12(20%), dfA17(9%), dfrA7(9%), and dfrA16(5%). The aadA were the second most abundant cassettes and their prevalence was: - aadA1(25%), aadA2(21%), and aadA5(14%). Other cassettes occurred in lower prevalence of below 5%. Prevalence of Tn21, ISEcp1, ISCR1 and IS26 was 22%, 10%, 15%, and 7% respectively. Majority of Tn21 containing integrons carried a complete set of transposition genes while class 2 integrons were borne on Tn7 transposon. The qnrA genes were detected in 34(3%) isolates while 19(1%) carried qnrB. All qnr genes were in MDR strains carrying integrons containing the ISCR1. Close to 88% of blaTEM-52 were linked to IS26 while ≥ 80% of blaCTX-Ms and bla CMYs were linked to ISEcp1. Only a few studies have identified a blaCTX-M-9 containing an ISEcp1 element as reported in this study. Multiple genetic elements, especially those borne on incIl, incFII, and incL/M plasmids, and their associated resistance genes were transferrable en bloc to E. coli strain J53 in mating experiments.
Conclusions
This is the first detailed study on the prevalence of selected elements implicated in evolution of resistance determinants in a large collection of clinical E. coli in Africa. Proliferation of such strains carrying multiple resistance elements is likely to compromise the use of affordable and available treatment options for majority of poor patients in Africa. There is therefore a need to monitor the spread of these highly resistant strains in developing countries through proper infection control and appropriate use of antimicrobials.
Similar content being viewed by others
Background
Recent studies conducted in Kenya show that a significant proportion of E. coli strains from clinical specimens exhibit a strong multi-drug resistance (MDR) phenotype [1, 2]. Fortunately, β-lactams, fluoroquinolones and aminoglycosides remain effective against a significant proportion of clinical E. coli strains in Kenya. However, recent studies have reported carriage of plasmid-borne aac(6')-lb-cr and qnr genes among β-lactamase producers [1, 2]. The qnr genes confer resistance to quinolones, while aac(6')-lb-cr confers reduced susceptibility to fluoroquinolones and aminoglycosides. Therefore, carbapenems remain some of the few alternative antimicrobials that are effective against strains harboring a combination of multiple β-lactamase (bla) genes and genes conferring broad-spectrum resistance to fluoroquinolones and aminoglycosides. Carbapenems may however not be readily available or affordable for many patients in Sub-Saharan Africa [3].
In a recent study, we reported carriage of integrons, IS elements, Tn21 and Tn7 in a collection of 27 E. coli strains obtained from hospitalised patients [1]. These strains also harbored conjugatively transferrable plasmids conferring resistance to β-lactams, fluoroquinolones, aminoglycosides and co-trimoxazole among other antimicrobials suggesting that genes encoding resistance to these antimicrobials are physically linked to each other. Carriage of physically linked elements, each containing a set of resistance genes, may increases the chances of en bloc horizontal transfer of multiple resistance determinants to susceptible strains. Carriage of multiple resistance elements may in turn confer unique advantages to the host and enable them survive a strong antimicrobial selection pressure especially in hospital settings [4].
Studies to determine the prevalence of resistance elements in a large collection of strains from Sub-Saharan Africa are still lacking. Furthermore, little is known on whether the genetic elements encountered among E. coli strains in this region are physically linked to each other. In this study, we determined the prevalence of integrons, ISEcp1, ISCR1, IS26 as well as transposons Tn21 and Tn7 in a collection of 1327 E. coli strains obtained from inpatient and outpatient populations seeking treatment in Kenyan hospitals during a 19-year period (1992–2011). We also determined genetic content of integrons and determined plasmid incompatibility groupings among strains exhibiting unique resistance phenotypes. Physical linkages among these elements and to bla genes were investigated using PCR methods. Similar analysis were done to determine if the aac(6')-lb-cr and qnr genes are physically linked to these elements.
Results
Antimicrobial susceptibility profiles
At least 25% of the 1327 isolates were resistant to expanded-spectrum β-lactams such as aztreonam (AZT), ceftriaxone (CRO), cefotaxime (CTX) and amoxicillin-clavulanic acid (AMC) combunation and to none-β-lactams such as streptomycin (S), nitrofurantoin (F), chloramphenicol (C), sulfamethoxazole (SUL), tetracyclines (TET) and trimethoprim (TRIM), Table 1. Resistance to a combination of two β-lactamase inhibitors, AMC and pipperacillin-tazobactam (TZP), was recorded in 22% of the isolates while 20% and 9% exhibited an ESBL- or an AmpC-like phenotype respectively, Table 2. A total of 106 strains were resistant to combinations of SUL, TRIM, ciprofloxacin (CIP), cefepime (FEP), gentamicin (CN), cefoxitin (FOX) and TZP. These isolates were therefore identified as strains with the highest potential to limit therapeutic option in clinical settings. Imipenem (IMI), cefepime FEP and CIP were effective against ≥ 90% of isolates. Strains from urine were more likely to exhibit an MDR phenotype compared to those from stool (p:0.0001, CI:27.2 to 84.8, OR:42) or blood (p:0.0001, CI:12.8 to 30.8, OR:19.9). Similarly, MDR phenotypes were more common among strains from hospitalized patients than those from non-hospitalized patients (p:0.0001, CI: 4.0 to 6.6, OR:5.1).
Prevalence of integrons and integron cassettes
Class 1 integrons were detected in 35% of all isolates, 3 isolates carried class 2 integrons but none tested positive for class 3 integrons. The dfrA sub-types conferring resistance to TRIM and the aadA-type cassettes conferring resistance to aminoglycosides were the most common cassettes in class 1 and 2 integrons, Table 3. The prevalence of cassettes encoding resistance to trimethoprim was: - dfrA1 (28%), dfrA12 (20%), dfA17 (9%), dfrA7(9%), and dfrA16 (5%), while that of aadA cassettes conferring resistance to aminoglycosides was as follows: - aadA1 (25%), aadA2 (21%), and aadA5 (14%). Despite a relatively high prevalence of resistance to β-lactams, only blaOXA-1 was identified as an integron cassette. While aadA and dfrA types were detected in strains exhibiting resistance to between 2 and 8 classes of antimicrobials, dfrB, aadA5, blaOXA-1, aac(6’)-lb-cr, and arr2 were detected only in strains resistant to at least 6 different classes of antimicrobials. Majority (78%) of dfrA17 were detected in strains resistant to multiple generations of β-lactams.
The cmlA1 and aadA1/dfrA1 cassette arrays were only detected in integrons containing a 3’-CS. In contrast, at least 64% of aac(6')-lb-cr, dfrA12/aadA1/blaoxa1,orf5/dfrB/orfA, and aac(6')Ib/blaoxa-1/catB3/arr2 cassette arrays were detected in integrons lacking typical 3’-conserved sequences (3’-CS) that contains qacEΔ1 (a truncated gene encoding resistance to quaternary ammonium compounds, and sul1 encoding resistance to sulfonamides). All the three class 2 integrons contained an identical cassette array comprising dfrA1-sat2-aadA1.
Prevalence of Tn21, Tn7and IS elements
The prevalence of Tn21 was 22% while Tn7 was detected in 3 isolates that also carried class 2 integrons. Prevalence of ISEcp1, ISCR1 and IS26 was 10%, 15%, and 7% respectively. A high proportion (≥ 60%) of isolates containing the IS elements and integrons were MDR (resistant to at least 3 different classes of antimicrobials), Table 4. Isolates carrying multiple elements were more likely to exhibit an MDR phenotype than those lacking such elements (p:0.0001, CI:549.5 to 2419.6, OR:1153) and isolates from urine were more likely to harbor multiple elements compared to those from blood (p:0.0001, CI:3.1 to 5.5, OR:4.1) or those from stool (p:0.0008, CI:1.2 to 2.0, OR:1.6). Although integrons, IS elements and Tn21 were detected in isolates from all specimen-types, a high proportion (69%) of these elements were detected among strains from urine of hospitalized patients.
Physical linkage amongst genetic elements
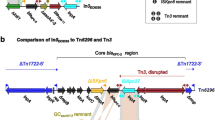
Figure 1 illustrates the strategy used for interrogation for physical linkages amongst genetic elements while Figure 2 illustrates some of the genetic associations identified in this study. Majority (69%) of integrons containing 3’-CS were physically linked to the Tn21 transposon while 75% of those containing a sul3 gene at the 3’-terminal were linked to IS26. This element was also linked to 80% of integrons lacking the 3’-CS, Table 5. Forty (40) isolates contained class 1 integrons linked to a single IS26 upstream the 5’-CS while in 12 isolates the integrons was flanked by two IS26 elements. All ISCR1 were detected only in MDR strains and were flanked by a pair of class 1 integron 3’-CS. Close to 94% of Tn21 that were linked to an integron contained a complete set of transposition genes (tnpA, tnpR and tnpM) while 89% of Tn21 with an incomplete set of these genes did not contain an integron, Table 6. All the three class 2 integrons were physically linked to Tn7.
Schematic diagram showing some of the strategies for screening for various genetic elements and for interrogation between these elements and resistance genes. The targets of each primer and the direction of PCR amplification is shown using arrows. PCRs were done both in the 5’ and in the 3’ orientation for each pair of genes tested. A: The strategy used for detection and characterization of class 1 integrons. B: The strategy used for detection and characterization of class 2 integrons and their physical linkage to Tn7. C: An example of the strategy used for analysis of physical linkages between class 1 integrons and Tn21 and to IS26. The primer positions for screening of Tn21 transposition genes. D and E: An example of the strategy used for analysis for physical linkages between integrons, ISCR1 and bla genes. F: An example of the strategy used for analysis for physical linkages between integrons, ISEcp1, IS26 and bla genes. These illustrations are based on PCR mapping data and not sequencing. Therefore, the sizes of each gene and the distances between any two genes are not drawn to scale.
Schematic diagram illustrating examples of physical linkages amongst genetic elements and selected genes. 1a-1f: An example of physical linkages between bla genes and multiple genetic elements such as integrons, ISEcp1, and IS26. 2a-2b: An example of physical linkages between bla genes and ISEcp1. 3a-3d: An example of physical linkages between integrons and other genetic elements (such as the ISCR1 element) that are in turn linked to bla genes and (fluoro)quinolone resistant genes. 4a-4c: An example of physical linkages between Tn21 and integrons that are in turn be linked to IS elements. These illustrations are based on PCR mapping data and not sequencing. Therefore, the sizes of each gene and the distances between any two genes are not drawn to scale.
Physical linkages between resistance genes and genetic elements
Figure 2 illustrates selected examples of physical linkages between bla genes and different genetic elements. Over 40% of isolates carrying blaTEM-52, blaSHV-5 or blaCTX-M-14 were physically linked to the IS26, Table 7. The ISEcp1 was the most common IS element associated with blaCTX-M-14,bla CTX-M −15 and blaCMY-2. One isolate contained a blaCTX-M-9 linked to this element. In all cases, the ISEcp1 was detected upstream the bla gene, Figure 2.
Thirty seven (88%) of the 42 aac(6’)-lb-cr were borne on integrons containing the ISCR1 while 55% were borne on integrons linked to the IS26. Twenty four (71%) of the 34 isolates carrying a qnrA gene were resistant to nalidixic acid but not to ciprofloxacin while the other 10 isolates carrying this gene and 19 carrying the qnrB subtype were resistant to both antimicrobials, Table 8. None of the isolates tested positive for qnrS. Majority (87%) of qnr genes were physically linked to either integron-associated ISCR1 or the IS26. All Isolates carrying aac(6’)-lb-cr or the qnr genes contained multiple genetic elements and were all MDR.
Conjugative plasmids mediate en bloctransfer of multiple elements and resistance genes
Multiple resistance genes and genetic elements associated with them were transferred en bloc to E. coli J53 in mating experiments, Table 9. Majority of such transferred were mediated by plasmids containing I1, L/M, XI, HI2 and the F-type replicons. These experiments further revealed that genes conferring resistance to tetracylines and chloramphenicol were also harbored in the same plasmids encoding resistance to β-lactams, (fluoro)quinolones and aminoglycosides. However, various gene combinations that had been determined to be physically linked using PCR could not be transferred in conjugation experiments using media containing different combinations of antimicrobials.
Discussion
The current study shows that a significant proportion of clinical E. coli strains in Kenyan are resistant to important classes of antimicrobials such as β-lactams, fluoroquinolones and aminoglycosides. These results are in agreement with those published before [1, 3, 5]. These MDR strains were however susceptible to carbapenems. It is easy (although illegal) to purchase antimicrobials in Kenya without prescriptions or with prescriptions not backed by laboratory investigations [6]. We hypothesize that such practices may directly or indirectly lead to emergence of highly resistant strains.
A high prevalence of MDR strains from urine and all specimens from hospitalized patients may reflects a corresponding heavy use of antimicrobials among this category of patients as reported in past studies [7, 8]. Majority of resistances encountered in hospital isolates were also encountered in community settings probably because patients are often discharged from hospitals as soon as their conditions improve, even before they complete their treatment regiments (our unpublished observations). It is therefore possible that hospital strains find their way into community settings and vis versa. However, we do not rule out the possibility that some MDR phenotypes may arise in community settings.
The high prevalence of class 1 integrons may partially be due to their association with the Tn21 that contain a complete set of transposition genes. Past studies show that dfrA7 and dfrA1 cassettes associated with Tn21-borne integrons are the most prevalent dfrA-subtypes in Central, North and Western Africa [9–12]. In this study however, the prevalence of dfrA7 was much lower than that of dfrA1, dfrA12 and dfA17 in that order. The class 2 integron dfrA1/sat2/aadA1 array reported in this study is globally distributed [13]. Our results may therefore reflect regional differences or similarities in distribution of integron cassette arrays. Such differences may arise from unique antimicrobial-use patterns in different countries. This study also demonstrates an apparent correlation between carriage of dfrA17 and resistance to multiple β-lactams as has been reported in Tunisia [12, 14] and from Northern Kenya among isolates from dog, cat and human specimens [5]. The reasons behind these correlations are yet to be elucidated. Carriage of different dfrA sub-types in our isolates and carriage of multiple integron-associated sul genes (sul1 and sul3) in the same isolate possibly correlates to heavy usage of sulfonamides and trimethoprim in Kenya for treatment of different infections and as prophylaxis against opportunistic infections among people with HIV/AIDS [15–17].
Some integrons, especially those lacking the 3’-CS and those containing a sul3 at the 3’-end, were linked to the IS26 possibly because this element mediates deletion of 3’-CS in class 1 integrons 3’- terminal [18, 19]. Similar results have been published in Australia, Spain and Nigeria [11, 12, 18, 19]. Our data further suggest that strains carrying IS26-associated integrons are highly MDR probably because the IS26 is also linked to other non-integron genes such as β-lactamases.
Most β-lactamases, particularly those encoding CTX-M-14 and −15 and CMY-2, were physically linked to ISEcp1. Similar reports have been published in Tunisian [20, 21] but no ISEcp1 was detected upstream the bla-CTX-M-1 among our isolates as reported in a related study from the same country [22]. In one isolate, this element was found upstream the blaCTX-M-9. Reports of ISEcp1-blaCTX-M-9 linkages are rare but such linkages have been reported in Klebsiella pneumoniae isolates in Taiwan [23]. Majority of bla TEM genes, blaTEM-52 in particular, were physically linked to the IS26 as reported in Belgium and Germany [24, 25]. Taken together, these results suggest that most bla genes in our isolates are in similar genetic environments as those reported globally but the genetic environment of blaCTX-M-9 and blaCTX-M-1 in our isolates appears to be different from those reported globally.
Our results further demonstrated that most bla genes are distantly linked to elements that are in turn linked to other resistance genes such as aac(6’)-lb-cr and qnr. Similar reports have been published in Tunisia [20, 21] and in Nigeria [11]. ISEcp1, IS26 and ISCR1 are known to mediate transposition and/or expression of multiple resistance genes in their close proximity [26–31]. Carriage of such multiple elements, each carrying a set of resistance genes may be responsible for the observed co-resistance to multiple antimicrobials among our isolates.
Conjugation experiments confirmed that multiple elements were borne on narrow host-range plasmids such as IncFII, IncH12 or on broad host-range plasmids such as IncL/M. The type of conjugative plasmids in our isolates (especially those carrying plasmids containing incF-type, incHI2 and incI1 incL/M replicons) were shown to confer resistances similar to those in strains from Tunisia, [32] and from two other studies conducted in Kenya [1, 5]. We hypothesis that plasmids of different incompatibility groups have acquired similar or identical sets of resistance genes and this acquisition is mediated by genetic elements such as those investigated in this study. Therefore, there is a possibility that such elements act as genetic shuttles between plasmids of different incompatibility grouping. The similarities and differences in genetic environments of bla, aac (6’)-lb-cr and qnr genes reported in this study may reflect a difference in transposition activities of such elements. We further hypothesize that differences in antibiotic use patterns in different regions influence the transposition activity of such elements.
Conclusions
This study reports carriage of multiple genetic elements in MDR E. coli strains and their association with selected resistance genes. Strains carrying such elements are likely to be well adapted to survive deleterious effects of combined antimicrobial therapy. Furthermore, such MDR strains have a potential to increase morbidity and mortality among patients. It is therefore important to launch surveillance programs and to put up measures to curtail the spread of these highly resistant strains. There is also a need to compare the genomes of strains encountered in Africa with those from other parts of the world.
Methods
Isolates
The 1327 non-duplicate isolates were obtained sequentially from 13 healthcare facilities in Kenya between 1992 and 2011 (19-year period) from 654 hospitalized and 673 non-hospitalized patients. These isolates comprised of 451 strains from patients with urethral tract infections (UTI) and those with urinary catheters while 371 were from blood of patients with septicemia. Another 505 strains were from fecal specimens of patients with loose stool, watery and bloody diarrhea. Only one isolate per specimen per patient was included for further analysis. Among the isolates investigated in this study, 912 had been analyzed for bla genes in a a past study [3] while 27 had been analyzed for selected genetic elements [1]. Ethical clearance to carry out this study was obtained from the KEMRI/National Ethics Committee (approval number SSC No. 1177).
Antimicrobial susceptibility profiles
Susceptibility profiles for all isolates were determined using antibiotic discs (Cypress diagnostics, Langdorp, Belgium) on Mueller Hinton agar (Oxoid, Louis, Mo. USA) using the Laboratory Standards Institute guidelines (CLSI) [33].
Detection of genetic elements
Figure 1 illustrates the strategy used for detection and characterization of integrons and transposons. Detection of class 1, 2 and 3 and determination of carriage of 3’-conserved sequences (3’-CS) in class 1 integrons was done as described before [34, 35]. Class 1 integron variable cassette region (VCR), the region in which the resistance gene cassettes are integrated, was amplified as previously described by Dalsgaard et al.[35] while that of class 2 integrons was amplified as described by White et al.[36]. The VCRs of integrons lacking the typical 3’-CS was determined using a PCR walking strategy published before [37]. Identification of integron cassette identity was done using a combination of restriction fragment length polymorphism (RFLP), sequencing and published bioinformatics tools [38, 39]. Detection of the ISEcp1, ISCR1, Tn21 and Tn7 elements was done as described in published studies [34, 35]. Analysis for Tn21 transposition genes:- tnpA, tnpR and tnpM genes was done as previously described by Pearson et al.[40]. The primers used in this study are presented in Table 10.
Detection of aac(6’)-lb-cr and qnrgenes
Screening for aac(6′)-Ib-cr gene that confers cross-resistance to fluoroquinolones and aminoglycoside was done using a combination of PCR, RFLP and sequencing as described by Park et al.[41]. The isolates were also screened for genes conferring resistance to quinolones: - qnrA, qnrB and qnrS using PCR and sequencing strategies previously described by Wu et al.[42].
Interrogation for physical linkages between genetic elements and resistance genes
Physical linkages between integron and the transposons were determined using a combination of published primers targeting 5’-conserved sequences (5’-CS) of class 1 integrons and those targeting the tnpM of Tn2 or those specific for tnpA7 of Tn7, Figure 1. A combination of primers targeting IS elements and those targeting the 5’-CS or the 3’-termini of integrons were used for interrogation for physical linkages between integrons and IS elements. A combination of primers specific for various genetic elements and consensus primers for bla SHV or blaTEM,[43, 44], bla CTX-M [45], bla CMY [46] and bla OXA [47, 48] were used for determination of physical linkages between bla genes and different genetic elements. Primers for aac(6’)-lb-cr and qnr genes were used in combination with those for different genetic elements to analyze for their physical association. A long-range polymerase [LongAmp® Taq DNA Polymerase, (New England Biolabs, USA)] was used in all reactions for physical linkages. A slow ramping rate of between 0.2°C/sec and 0.3°C/sec was set for the annealing step. The extension time was set at 72°C for 2 min and a final extension of 72°C for 15 min was carried out after 35–40 cycles of denaturation, annealing and extension.
Conjugation experiments
Conjugation experiments using sodium azide resistant E. coli strain J53 as the recipient were done as previous described [49]. Susceptibility to antimicrobials and determination of genetic element content of the transconjugants was determined using similar methods as those used for the corresponding donor strains. Plasmid incompatibility groupings were determined using the scheme of Carattoli et al.[50].
Statistical analysis
For the purpose of analysis, both intermediate and resistant results for antibiotic susceptibility testing were grouped together as “resistant”. Differences in proportion of isolates bearing different elements was analyzed using the Chi test (χ2) while the Fisher’s exact test was used for smaller sample sizes. The Odds Rations (OR) and the 95% confidence intervals (CIs) accompanying the χ2 tests were determined using the approximation of Woolf. The null hypothesis was rejected for values of p ≥ 0.05. Statistical analysis was performed using Statgraphics plus Version 5 (StatPoint Technologies, INC, Warrenton, VA, USA).
Authors’ information
JK and SK are research scientists at the Kenya Medical Research Institute (KEMRI). BMG is Professor at the K.U.Leuven (Faculty of Bioscience Engineering) while PB is a Senior Research Scientist at the Veterinary and Agrochemical Research Centre (VAR).
References
Kiiru J, Kariuki S, Goddeeris BM, Revathi G, Maina TW, Ndegwa DW, Muyodi J, Butaye P: Escherichia coli strains from Kenyan patients carrying conjugatively transferable broad-spectrum beta-lactamase, qnr, aac(6')-Ib-cr and 16S rRNA methyltransferase genes. J Antimicrob Chemother. 2011, 66: 1639-1642. 10.1093/jac/dkr149.
Kariuki S, Revathi G, Corkill J, Kiiru J, Mwituria J, Mirza N, Hart CA: Escherichia coli from community-acquired urinary tract infections resistant to fluoroquinolones and extended-spectrum beta-lactams. J Infect Dev Ctries. 2007, 1: 257-262.
Kiiru J, Kariuki S, Goddeeris BM, Butaye P: Analysis of beta-lactamase phenotypes and carriage of selected beta-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol. 2012, 12: 155-10.1186/1471-2180-12-155.
Sabate M, Navarro F, Miro E, Campoy S, Mirelis B, Barbe J, Prats G: Novel complex sul1-type integron in Escherichia coli carrying bla(CTX-M-9). Antimicrob Agents Chemother. 2002, 46: 2656-2661. 10.1128/AAC.46.8.2656-2661.2002.
Albrechtova K, Dolejska M, Cizek A, Tausova D, Klimes J, Bebora L, Literak I: Dogs of nomadic pastoralists in northern Kenya are reservoirs of plasmid-mediated cephalosporin- and quinolone-resistant Escherichia coli, including pandemic clone B2-O25-ST131. Antimicrob Agents Chemother. 2012, 56: 4013-4017. 10.1128/AAC.05859-11.
Brooks JT, Shapiro RL, Kumar L, Wells JG, Phillips-Howard PA, Shi YP, Vulule JM, Hoekstra RM, Mintz E, Slutsker L: Epidemiology of sporadic bloody diarrhea in rural Western Kenya. Am J Trop Med Hyg. 2003, 68: 671-677.
Blango MG, Mulvey MA: Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother. 2010, 54: 1855-1863. 10.1128/AAC.00014-10.
Bejon P, Mwangi I, Ngetsa C, Mwarumba S, Berkley JA, Lowe BS, Maitland K, Marsh K, English M, Scott JA: Invasive Gram-negative bacilli are frequently resistant to standard antibiotics for children admitted to hospital in Kilifi, Kenya. J Antimicrob Chemother. 2005, 56: 232-235. 10.1093/jac/dki145.
Frank T, Gautier V, Talarmin A, Bercion R, Arlet G: Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). J Antimicrob Chemother. 2007, 59: 742-745. 10.1093/jac/dkl538.
Gassama A, Aidara-Kane A, Chainier D, Denis F, Ploy MC: Integron-associated antibiotic resistance in enteroaggregative and enteroinvasive Escherichia coli. Microb Drug Resist. 2004, 10: 27-30. 10.1089/107662904323047763.
Labar AS, Millman JS, Ruebush E, Opintan JA, Bishar RA, Aboderin AO, Newman MJ, Lamikanra A, Okeke IN: Regional dissemination of a trimethoprim-resistance gene cassette via a successful transposable element. PLoS One. 2012, 7: e38142-10.1371/journal.pone.0038142.
Dahmen S, Mansour W, Boujaafar N, Arlet G, Bouallegue O: Distribution of cotrimoxazole resistance genes associated with class 1 integrons in clinical isolates of Enterobacteriaceae in a university hospital in Tunisia. Microb Drug Resist. 2010, 16: 43-47. 10.1089/mdr.2009.0091.
Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, Grady M, Liebert C, Summers AO, White DG, Maurer JJ: Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother. 2001, 45 (3): 723-726. 10.1128/AAC.45.3.723-726.2001.
Ben SK, Jouini A, Ben SR, Somalo S, Saenz Y, Estepa V, Boudabous A, Torres C: Prevalence of broad-spectrum cephalosporin-resistant Escherichia coli isolates in food samples in Tunisia, and characterization of integrons and antimicrobial resistance mechanisms implicated. Int J Food Microbiol. 2010, 137: 281-286. 10.1016/j.ijfoodmicro.2009.12.003.
Grape M, Farra A, Kronvall G, Sundstrom L: Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. Clin Microbiol Infect. 2005, 11: 185-192. 10.1111/j.1469-0691.2004.01059.x.
White PA, McIver CJ, Rawlinson WD: Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001, 45: 2658-2661. 10.1128/AAC.45.9.2658-2661.2001.
Bennett PM: Integrons and gene cassettes: a genetic construction kit for bacteria. J Antimicrob Chemother. 1999, 43 (1): 1-4. 10.1093/jac/43.1.1.
Curiao T, Canton R, Garcillan-Barcia MP, de la Cruz F, Baquero F, Coque TM: Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-beta-lactamase-producing Escherichia coli clones from humans. Antimicrob Agents Chemother. 2011, 55: 2451-2457. 10.1128/AAC.01448-10.
Dawes FE, Kuzevski A, Bettelheim KA, Hornitzky MA, Djordjevic SP, Walker MJ: Distribution of class 1 integrons with IS26-mediated deletions in their 3'-conserved segments in Escherichia coli of human and animal origin. PLoS One. 2010, 5: e12754-10.1371/journal.pone.0012754.
Ben SK, Ben SR, Jouini A, Rachid S, Moussa L, Saenz Y, Estepa V, Somalo S, Boudabous A, Torres C: Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr Microbiol. 2011, 62: 1794-1801. 10.1007/s00284-011-9930-4.
Jouini A, Ben SK, Vinue L, Ruiz E, Saenz Y, Somalo S, Klibi N, Zarazaga M, Ben MM, Boudabous A, Torres C: Detection of unrelated Escherichia coli strains harboring genes of CTX-M-15, OXA-1, and AAC(6')-Ib-cr enzymes in a Tunisian hospital and characterization of their integrons and virulence factors. J Chemother. 2010, 22: 318-323.
Ben SR, Ben SK, Estepa V, Jouini A, Gharsa H, Klibi N, Saenz Y, Ruiz-Larrea F, Boudabous A, Torres C: Prevalence and characterisation of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates in healthy volunteers in Tunisia. Eur J Clin Microbiol Infect Dis. 2012, 31: 1511-1516. 10.1007/s10096-011-1471-z.
Ma L, Siu LK, Lu PL: Effect of spacer sequences between bla(CTX-M) and ISEcp1 on bla(CTX-M) expression. J Med Microbiol. 2011, 60: 1787-1792. 10.1099/jmm.0.033910-0.
Cullik A, Pfeifer Y, Prager R, von BH, Witte W: A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J Med Microbiol. 2010, 59: 580-587. 10.1099/jmm.0.016188-0.
Smet A, Van NF, Vandekerckhove TT, Martel A, Deforce D, Butaye P, Haesebrouck F: Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLoS One. 2010, 5: e11202-10.1371/journal.pone.0011202.
Tavakoli N, Comanducci A, Dodd HM, Lett MC, Albiger B, Bennett P: IS1294, a DNA element that transposes by RC transposition. Plasmid. 2000, 44: 66-84. 10.1006/plas.1999.1460.
Toleman MA, Bennett PM, Walsh TR: ISCR elements: novel gene-capturing systems of the 21st century?. Microbiol Mol Biol Rev. 2006, 70 (2): 296-316. 10.1128/MMBR.00048-05.
Bado I, Cordeiro NF, Robino L, Garcia-Fulgueiras V, Seija V, Bazet C, Gutkind G, Ayala JA, Vignoli R: Detection of class 1 and 2 integrons, extended-spectrum beta-lactamases and qnr alleles in enterobacterial isolates from the digestive tract of Intensive Care Unit inpatients. Int J Antimicrob Agents. 2010, 36: 453-458. 10.1016/j.ijantimicag.2010.06.042.
Xia R, Guo X, Zhang Y, Xu H: qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob Agents Chemother. 2010, 54: 3471-3474. 10.1128/AAC.01668-09.
Bonnet R: Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004, 48: 1-14. 10.1128/AAC.48.1.1-14.2004.
Poirel L, Lartigue MF, Decousser JW, Nordmann P: ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother. 2005, 49 (1): 447-450. 10.1128/AAC.49.1.447-450.2005.
Zucker JR, Lackritz EM, Ruebush TK, Hightower AW, Adungosi JE, Were JB, Metchock B, Patrick E, Campbell CC: Childhood mortality during and after hospitalization in western Kenya: effect of malaria treatment regimens. Am J Trop Med Hyg. 1996, 55: 655-660.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing. Sixteenth informational supplement M100-S16. 2006, Wayne, PA: Clinical and Laboratory Standards Institute
Shi L, Fujihara K, Sato T, Ito H, Garg P, Chakrabarty R, Ramamurthy T, Nair GB, Takeda Y, Yamasaki S: Distribution and characterization of integrons in various serogroups of Vibrio cholerae strains isolated from diarrhoeal patients between 1992 and 2000 in Kolkata, India. J Med Microbiol. 2006, 55: 575-583. 10.1099/jmm.0.46339-0.
Dalsgaard A, Forslund A, Serichantalergs O, Sandvang D: Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob Agents Chemother. 2000, 44: 1315-1321. 10.1128/AAC.44.5.1315-1321.2000.
White DG, Zhao S, Simjee S, Wagner DD, McDermott PF: Antimicrobial resistance of foodborne pathogens. Microbes Infect. 2002, 4: 405-412. 10.1016/S1286-4579(02)01554-X.
Saenz Y, Vinue L, Ruiz E, Somalo S, Martinez S, Rojo-Bezares B, Zarazaga M, Torres C: Class 1 integrons lacking qacEDelta1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet Microbiol. 2010, 144: 493-497. 10.1016/j.vetmic.2010.01.026.
Yu HS, Lee JC, Kang HY, Jeong YS, Lee EY, Choi CH, Tae SH, Lee YC, Seol SY, Cho DT: Prevalence of dfr genes associated with integrons and dissemination of dfrA17 among urinary isolates of Escherichia coli in Korea. J Antimicrob Chemother. 2004, 53: 445-450. 10.1093/jac/dkh097.
Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22: 4673-4680. 10.1093/nar/22.22.4673.
Pearson AJ, Bruce KD, Osborn AM, Ritchie DA, Strike P: Distribution of class II transposase and resolvase genes in soil bacteria and their association with mer genes. Appl Environ Microbiol. 1996, 62: 2961-2965.
Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC: Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006, 50 (11): 3953-3955. 10.1128/AAC.00915-06.
Wu JJ, Ko WC, Wu HM, Yan JJ: Prevalence of Qnr determinants among bloodstream isolates of Escherichia coli and Klebsiella pneumoniae in a Taiwanese hospital, 1999–2005. J Antimicrob Chemother. 2008, 61 (6): 1234-1239. 10.1093/jac/dkn111.
Arlet G, Rouveau M, Philippon A: Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol Lett. 1997, 152 (1): 163-167. 10.1016/S0378-1097(97)00196-1.
Arlet G, Brami G, Decre D, Flippo A, Gaillot O, Lagrange PH, Philippon A: Molecular characterisation by PCR-restriction fragment length polymorphism of TEM beta-lactamases. FEMS Microbiol Lett. 1995, 134: 203-208.
Lartigue MF, Poirel L, Nordmann P: Diversity of genetic environment of bla(CTX-M) genes. FEMS Microbiol Lett. 2004, 234 (2): 201-207. 10.1111/j.1574-6968.2004.tb09534.x.
Winokur PL, Brueggemann A, DeSalvo DL, Hoffmann L, Apley MD, Uhlenhopp EK, Pfaller MA, Doern GV: Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob Agents Chemother. 2000, 44: 2777-2783. 10.1128/AAC.44.10.2777-2783.2000.
Olesen I, Hasman H, Aarestrup FM: Prevalence of beta-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb Drug Resist. 2004, 10 (4): 334-340. 10.1089/mdr.2004.10.334.
Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM: beta-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother. 2005, 56: 115-121. 10.1093/jac/dki190.
Jeong JY, Yoon HJ, Kim ES, Lee Y, Choi SH, Kim NJ, Woo JH, Kim YS: Detection of qnr in clinical isolates of Escherichia coli from Korea. Antimicrob Agents Chemother. 2005, 49 (6): 2522-2524. 10.1128/AAC.49.6.2522-2524.2005.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ: Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005, 63 (3): 219-228. 10.1016/j.mimet.2005.03.018.
Acknowledgements
The authors would like to thank staff and students attached to the CMR-WT unit lab at KEMRI and staff members of Bacteriology unit at VAR-Belgium. This work was supported by a PhD scholarship grant from the Vlaamse Interuniversitaire Raad (VLIR), Belgium (Grant number BBTP2007-0009-1086). This work is published with permission from the Director, KEMRI.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
None of the authors have competing interests.
Authors’ contributions
JK designed the study, carried out the experiments and wrote the manuscript. SK, BM and PB participated in manuscript write-up and review. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kiiru, J., Butaye, P., Goddeeris, B.M. et al. Analysis for prevalence and physical linkages amongst integrons, ISEcp1, ISCR1, Tn21 and Tn7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011). BMC Microbiol 13, 109 (2013). https://doi.org/10.1186/1471-2180-13-109
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-13-109