Abstract
Background
The traditional strategy to map QTL is to use linkage analysis employing a limited number of markers. These analyses report wide QTL confidence intervals, making very difficult to identify the gene and polymorphisms underlying the QTL effects. The arrival of genome-wide panels of SNPs makes available thousands of markers increasing the information content and therefore the likelihood of detecting and fine mapping QTL regions. The aims of the current study are to confirm previous QTL regions for growth and body composition traits in different generations of an Iberian x Landrace intercross (IBMAP) and especially identify new ones with narrow confidence intervals by employing the PorcineSNP60 BeadChip in linkage analyses.
Results
Three generations (F3, Backcross 1 and Backcross 2) of the IBMAP and their related animals were genotyped with PorcineSNP60 BeadChip. A total of 8,417 SNPs equidistantly distributed across autosomes were selected after filtering by quality, position and frequency to perform the QTL scan. The joint and separate analyses of the different IBMAP generations allowed confirming QTL regions previously identified in chromosomes 4 and 6 as well as new ones mainly for backfat thickness in chromosomes 4, 5, 11, 14 and 17 and shoulder weight in chromosomes 1, 2, 9 and 13; and many other to the chromosome-wide signification level. In addition, most of the detected QTLs displayed narrow confidence intervals, making easier the selection of positional candidate genes.
Conclusions
The use of higher density of markers has allowed to confirm results obtained in previous QTL scans carried out with microsatellites. Moreover several new QTL regions have been now identified in regions probably not covered by markers in previous scans, most of these QTLs displayed narrow confidence intervals. Finally, prominent putative biological and positional candidate genes underlying those QTL effects are listed based on recent porcine genome annotation.
Similar content being viewed by others
Background
Hundreds of QTLs have been identified in porcine species (pigQTL database), but there are still relatively few examples for which the mutations that underlie mapped QTLs have been identified [1–4]. The traditional strategy to map QTLs has been to use linkage analysis employing a limited number of microsatellite markers. These analyses usually mapped the QTLs to large intervals, 20 cM or more, which made it difficult to identify the underlying gene and mutation. The success in the positional cloning of these QTLs in domestic animals has been hampered by the absence of high-resolution linkage maps (several markers per cM) [5]. However, the arrival of genome-wide panels of SNPs makes available thousands of markers per chromosome increasing the information content and therefore the likelihood of detecting and fine map QTL regions [5, 6].
The Iberian x Landrace experimental cross (IBMAP) was developed to detect QTLs for several economic traits, including growth, fatness and carcass composition [7]. The whole genome QTL scan, carried out in the F2 population using 92 microsatellite markers covering the 18 autosomes, allowed to detect three significant QTLs in SSC2, SSC4 and SSC6 [8]. Subsequent studies and even obtaining new IBMAP generations [9] have delved into knowledge of these regions. Several candidate genes, such as LEPR, MTTP and FABP5, have been analyzed reporting some successful results [10–16].
Various studies have shown the utility of high-density SNP panels for linkage analyses by providing a greater information content in comparison to microsatellites [6, 17–19]. In the present study, we employed the porcine high density SNP panel, PorcineSNP60 BeadChip (Illumina), to carry out a genome QTL scan based on linkage mapping analyses using three of the generations of the IBMAP experimental population. The objective is to confirm previous QTL regions and especially identify new ones with narrow confidence intervals.
Methods
Animals and Phenotypic records
The animals and phenotypic information used in the current study belong to a F3 generation and two different backcrosses of the IBMAP experimental population [9, 12]. The IBMAP F1 generation was obtained from three Iberian Guadyerbas boars and 30 Landrace sows. Six F1 boars and 73 F1sows were parents of 577 F2 pigs. Five F1 boars were mated with 25 Landrace sows obtaining 160 backcrossed animals (BC1). In addition three of the F2 boars were mated with 15 F2 sows obtaining 68 animals of the F3 generation, and finally other four F2 boars were mated with 22 Landrace sows obtaining 79 backcrossed animals (BC2). Phenotypic records used in the analyses (Table 1) included the body weight (BW) measured at 150 days of mean age (BW150), and two backfat thickness measures, one at the level of the fourth rib at 4 cm of the midline using ultrasounds at 75 kg of mean weight (BFT75) and the other taken with a rule at slaughter (BFTS). Additionally, intramuscular fat content (IMF) measured by NIRS in longissimus dorsi samples and weights of primary cuts (hams, HW, shoulders, SW and loin bone-in, LBW) were also registered at slaughter.
All animal procedures were carried out according to Spanish and European animal experimentation ethics law and approved by the institutional animal ethics committee of IRTA.
SNP data
The 86 F3, 79 BC1 and 160 BC2 pigs, and their related animals from F2, F1 and F0 generations, 416 pigs in total, were genotyped with the PorcineSNP60 BeadChip (Illumina, Inc.), designed by Ramos et al. [20], using the Infinium HD Assay Ultra protocol (Illumina, Inc.). GenomeStudio software (Illumina, Inc.) was employed for visualize, edit and filter the genotyping data. Raw individual data had high-genotyping quality (call rate >0.99). The SNPs filtering was carried in our previous study [21]. Briefly, those SNPs with GenTrain Score lower than 0.85, non-Mendelian inheritance, minor allele frequency less than 0.15, located in sex chromosomes, unmapped in the Sscrofa10 assembly or showing position errors in the linkage mapping were discarded using Plink software [22]. A total of 28,633 SNPs were retained in the dataset after quality control and filtering. In addition, a selection of the most informative SNPs was carried out based in their genetic distance according to the linkage maps generated in our previous study [21]. When the genetic distance among contiguous SNPs was 0, one of them was retained as representative of the linkage group for further analyses.
QTL scan
The linkage maps used for the QTL scan were obtained in Muñoz et al. [21] previous study. A joint QTL scan was performed in all BC1, BC2 and F3 animals. Moreover, two separate analyses were carried out in the BC1 and in the F3 + BC2 animals in agreement with their different parental boar origin. The QTL scans were performed with the following basic model:
where y ijk is the ijkth observation for the analyzed trait, Si and Bj are the systematic effects for sex (male or female) and batch (eight levels in the whole analysis, three or five levels in the analysis of BC1 or F3 + BC2 pigs), uk is the random polygenic effect of the kth individual, xk is a covariable (individual age, body or carcass weight in different analyses) and b its respective slope, a is the QTL additive effect; P ak is the additive coefficient calculated as P ak = Pr(QQ) - Pr(qq), the probability of the kth individual being homozygous for alleles of Iberian origin minus the probability of being homozygous of alleles of Landrace origin and eijk is the random residual. The infinitesimal genetic effect was treated as random, with covariance Aσu2A being the numerator relationship matrix. A single residual variance is assumed for all generations (F3, BC1 and BC2). A similar model fitting different QTL effects was used for performing complementary analyses to test the hypothesis of two QTLs mapping in different positions of the same chromosome and with effects a1 and a2 on the same trait:
Finally, joint analyses for two traits were performed to test possible pleiotropic effects of some QTL. The used model was equivalent to the basic, but here the (co)variances of the infinitesimal genetic effects are A ⊗ , where ⊗ denotes the Kronecker product and the subindices y and z correspond to the traits.
Likelihood ratio tests (LRT) were calculated comparing the full model and a reduced model without the corresponding QTL effect. The nominal P-values were calculated assuming a χ2 distribution of the LRT with the degrees of freedom given by the difference between the number of estimated parameters in the reduced and full models. Taking the nominal P-values resulting from the simultaneous testing, their q-values were inferred using QVALUE software [23]. The cut-off of significant QTL at the genome and chromosome level was set at q-value < 0.10. The confidence intervals (CI) were calculated at 95 % following Mangin et al. [24].
Gene annotation
The physical positions of the SNPs were conducted following Sscrofa10.2 genome annotation. The SNP framing the QTL confidence intervals were used to explore gene contain in pig genome assembly 10.2. Gene annotations were retrieved from Gbrowse (http://www.animalgenome.org/cgi-bin/gbrowse/pig10/).
Association analyses
Complementary association analyses were performed for specific SNPs (and haplotypes) mapped within candidate genes and included in the porcine chip. Candidate genes were identified based on their position within the QTL intervals and their functional relation with the analyzed traits. By the comparison of the SNP position with the gene position, both following Sscrofa10.2, SNPs within the candidate gene were identified and association analyses were conducted. Haplotypes were determined using Haploview software [25].
The analyses were carried out using the standard animal model:
where λk is the vector that includes an indicative variable related with the number of copies of one of the SNP or haplotype alleles, which takes 1 or -1values when the kth animal was homozygous for each allele or 0 if the animal was heterozygous; g represented the additive effect of the SNP or haplotype.
All the statistical analyses were performed using the Qxpak v.5.1 software [26].
Results
A total of 8,417 SNPs evenly spaced were used for the analyses. The mean distance between SNPs ranged from 0.18 cM in SSC11 to 0.33 cM in SSC6 (Table 2). The QTL scan has allowed to confirm QTL regions previously identified in the IBMAP population as well as identify new ones (Table 3) and many others at chromosome-wide significant level that are considered as suggestive (Additional file 1: Table S1).
The joint scan of both populations (BC1, F3 + BC2) revealed QTL regions in ten of the 18 autosomes (Table 3 and Additional file 1: Table S1). Six of which were significant to the genome-wide level: three QTLs for BFT75 in SSC4, SSC11 and SSC17, two for SW in SSC1 and SSC4 and one for BLW in SSC4 (Table 3). A complementary analysis was carried out in order to test possible pleiotropic effects of the SSC4 QTL for SW and BLW (Table 3). The results showed a significant pleitropic QTL (P-value = 5.7 x 10-6) at 60 cM with additive effects on these traits (-0.24 ± 0.05 kg and -0.28 ± 0.07 kg, respectively).
The QTL detection analyses carried out in the BC1 generation revealed QTL regions in 14 of the 18 porcine autosomes (Tables 3 and Additional file 1: Table S1), four of which were significant to the genome-threshold level. These genome–wide QTLs were identified in SSC4, SSC11, SSC14 and SSC17 for BFT75 trait (Table 3).
The QTL scan in the F3 + BC2 generations showed QTL regions in 11 of the 18 porcine autosomes (Tables 3 and Additional file 1: Table S1). Ten of these were significant to the genome-wide level: for BFTS in SSC4, SSC5, SSC6 and SSC14, for SW in SSC2, SSC4, SSC6, SSC9 and SSC13 and for BLW in SSC2 (Table 3). A complementary analysis was carried out in order to test pleiotropic effects of the SSC2 QTL for SW and BLW (Table 3). The results showed a significant pleitropic QTL (P-value = 1.1 x 10-4) at 116 cM with additive effects on these traits (0.40 ± 0.11 kg and 0.74 ± 0.16 kg, respectively).
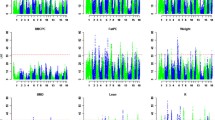
The separate analyses, using the same set of SNPs markers, evidenced differences between the populations. Examples of these differences are shown in Figure 1. The joint analyses allowed to capture some of the QTLs identified in the separates analysis (in SSC4, SSC11 and SSC17 for BFT75). Nevertheless, a QTL in SSC1 for SW reached the genome-wide significance in the joint analysis but not in the separate ones (Figure 2).
Discussion
The genome wide association study (GWAS) is the approach widely used for the analysis of high density SNP data. In the present study a classical QTL scan, based on the parent line origin assuming alternative alleles fixed in each of the parental populations, has been considered appropriate for the QTL detection analysis in agreement with the experimental design. The QTL scan using this high density panel of 8,417 SNPs has allowed the confirmation of QTL regions previously identified in the IBMAP population. Moreover, new QTLs have been detected, despite using a limited number of animal data, in regions probably not covered by the limited number of microsatellite markers used in previous studies.
Two different QTL analyses were carried out, a joint QTL scan and two separate analyses in agreement with the different parental boar origin of the generations. The separate analyses evidenced the differences between populations regarding the expected QTL genotypes and the random sampling of the QTL alleles in F1 and F2 boars. While only Qq and qq genotypes, coming from the F1 boars, are expected for the QTLs in the BC1 animals, the three possible QTL genotypes (QQ, Qq and qq, coming from the F2 boars) are possible in BC2 and F3 pigs. In addition, the F2 boars used for F3 and BC2 were selected conditioned on their potential genotypes for different QTL (mainly the QTL for growth and fatness in SSC4 and SSC6); however no selection could be done for the F1 boars used for BC1. These differences between populations are reflected in the results obtained. The joint analyses allowed to capture some of the QTLs identified in the separates analysis (in SSC4, SSC11 and SSC17 for BFT75) but not most of them. Nevertheless, other QTL, the one detected in SSC1 for SW, reached the genome-wide significance in the joint analysis but not in the separate one, indicating a gain of detection power with the increase of the record number for this QTL.
The most significant QTL region identified in the present study corresponded to the detected in SSC4 for BFT75, SW and BLW in the joint analysis. The likelihood profiles, shown as –log10(P-value), showed the presence of at least two QTL regions in SSC4, with a maximum located around 53-60 cM position for SW and BLW and around 104 cM for BFT75 (Figure 3). The first QTL region as well as the effects agree with one of the most relevant QTL for growth and fatness described in the IBMAP material for growth and fatness [8, 9], overlapping the known FAT1 region [27, 28]. Moreover, a complementary analysis revealed that this first QTL region presented pleotropic effects on SW, BLW and BFTS. However, the second QTL region, around 104 cM affecting BFT75 has been identified for the first time in the IBMAP material, but it has been already described in other populations (Table 4). In addition, the QTL significant profile of SSC4 scan for BFT75 in the BC1 may indicate another potential QTL region around 75 cM (Figure 3). Nevertheless, a complementary analysis employing a model with two QTL vs one single QTL did not allow to detect this possible secondary QTL. Another of the most relevant QTL regions for growth and fatness previously identified in the IBMAP experimental population was located around LEPR gene in SSC6 [10]. In the present analyses, this QTL has also been detected but only in the F3 + BC2 population (Figure 1). The QTL effects agree with the previously described for backfat thickness and shoulder weight. The Q Iberian allele led to an increase of the backfat and a decrease of the shoulder weight. This QTL could not be detected in the BC1, probably due to the lack of QTN segregation in this animal material. In fact, a previous study of the putative causal mutation of this QTL, LEPR c.1987 C > T, on this BC1 material, could not reveal significant associations due to the scarcity of some genotypes [13]. The remaining significant genome-wide QTL regions identified in the porcine chromosomes 1, 2, 5, 9, 11, 13, 14 and 17 in one or both populations have been previously associated with growth, fatness and conformation traits in the porcine species (Table 4). However, they were not detected in previous IBMAP scans, except the QTL in SSC9 for SW and in SSC14 for BFT but at suggestive signification level [8]. No genome-wide QTLs could be detected for W150d, IMF and HW traits, probably due to the limited size of the analyzed data, as several ones were detected to chromosome-wide significance level (Additional file 1: Table S1).
Most of the significant QTL regions identified in the present study displayed CI shorter than 5 cM (Table 3), which should facilitate the identification of suitable candidate genes. Thanks to the huge effort of Swine Genome Sequencing Consortium on porcine genome assembly and SNP annotation, a refined search of positional candidate genes could be carried out using Gbrowse tool (Table 5). It should be noted that porcine gene annotation is still scarce, several genes are projected but not annotated; nevertheless gene position within the QTL is more likely to be correct that those inferred from comparative humane-porcine mapping used in previous works. In addition, many annotations discrepancies exist between databases, most likely due to mapping differences between porcine genome versions. In the current study we have mainly used, assuming as more reliable, the latest Sscrofa10.2 genome version for gene annotation. The following paragraphs expose a discussion of the positional and biological putative candidate genes for each of the genome-wide significant QTL regions, according to current porcine gene annotation and available biological information.
The CI (102-104 cM, 232.7-240.4 Mb) of the SSC1 QTL for SW includes 11 protein-coding genes, however only two are annotated to known genes (Table 5). Although no study in porcine has been focused on PTPRD gene, studies in human suggest that PTPRD gene could play a relevant role in glucose homeostasis and insulin sensitivity [49].
Within the pleiotropic CI (112-117 cM, 150.9-158.3 Mb) of SSC2 QTL for SW and BLW, there are 63 protein-coding genes, 19 out of them are annotated to known genes (Table 5). Among these, PPARGC1B constitutes a strong candidate, although it has never been studied in porcine species. PPARGC1B belongs to the PGC-1 family, which act as coactivators in the dysregulation in diseases such as diabetes, obesity and cardiomyopathy in humans [50].
As it was mentioned before, the SSC4 QTL for fatness and conformation traits was identified in previous IBMAP scans [7, 8]. In addition, subsequent studies have aimed to deepen the knowledge of this region and some candidate genes (Figure 3) have been analyzed reporting different results [15, 16, 51–53]. In the present study, apart from that region around 60 cM, another QTL region for BFT75 has been identified around 104 cM position. Within the CI of this second QTL (102-109 cM, 129.7-134.4 Mb) there are 18 protein-coding genes, 12 out of them are annotated to known genes (Table 5), highlighting the AGL and VCAM1 genes as powerful biological candidates underlying the QTL effects. Han et al. [54] study revealed associations of an indel polymorphism in the AGL gene with growth, fatness and carcass traits in an F2 population crossbred Landrace and Jeju (Korea) Black pigs. Recently, Fontanessi et al. [55] study revealed associations of one SNP in VCAM1 gene with backfat thickness in Italian Large White pigs.
The SSC5 QTL for BFTS showed the largest CI (71-88 cM, 69.5-83.7 Mb), including more than 100 protein-coding genes, 34 of which are annotated to known genes (Table 5). Among the long list of putative candidates, ADIPOR2 and VDR genes highlight as powerful biological candidates, although they have never been studied as candidate gene for fatness in porcine species. The ADIPOR2 mediates the increased AMPK and PPAR-alpha ligand activities, as well as fatty acid oxidation and glucose uptake by adiponectin [56]. Human studies suggest that VDR may function as a determinant of muscle strength, fat mass and body weight [57].
The LEPR gene is the most powerful candidate underlying the QTL for fatness and conformation traits mapped in SSC6 in the F3 + BC2 generation. In fact, a highly significant association of a polymorphism located in exon 14, LEPR c. 1987 C > T, with growth and fatness has been previously found in several generations of the IBMAP population [11, 12]. These effects have been also confirmed in other porcine populations [58–61]. Moreover, functional studies have revealed differences in the LEPR mRNA expression levels in hypothalamus conditional on LEPR c.1987 C > T genotype [13] in agreement with the potential causal effect of this QTL on growth and fatness.
The CI (106-113 cM, 121.0-127.1 Mb) of the SSC9 QTL for SW includes 44 protein-coding genes, 13 out of them are annotated to known genes (Table 5). Among the potential list of candidates, the MYOG gene plays an essential role in the development and differentiation of muscle. Moreover, studies in porcine species have investigated the associations of MYOG polymorphisms with carcass composition and meat quality in pigs evidencing significant associations [62, 63]. Also, FASL gene has been implicated in skeletal myogenesis [64].
The SSC11 QTL for BFT75 showed one of the shortest CI (25-27 cM, 26.4-27.9 Mb). Within this region only nine protein-coding genes are projected, three of which are annotated to known genes (Table 5), however there is not a feasible candidate as the biological function of these genes have not been elucidated yet.
The CI (109-113 cM, 149.5-153.6 Mb) of SSC14 QTL for live backfat deposition and at slaughter includes 43 protein-coding genes. The CYP2E1 gene appears among the eight annotated to known genes. The CYP2E1 has been widely studied in pigs regarding boar taint [65–67], however, its relation to porcine lipid metabolism and fatness has never been explored, even if its key role in obesity and insulin resistance phenotypes has been showed in rodents and humans [68, 69].
The SSC13 QTL for SW showed a CI of 7 cM (47-53 cM, 83.4-91.0 Mb). Within this interval 64 projected protein-coding genes are mapped, 25 of which are already annotated to know genes (Table 5). Among them, NCK1 gene is found as a functional candidate. This gene encodes for a protein implicated in regulating the unfolded protein response, which secondary to obesity impairs glucose homeostasis and insulin actions [70].
Finally, the CI (56-61 cM, 39.6-42.1 Mb) of SSC17 QTL for BFT75 contains 64 coding-protein projected genes, 28 of which are annotated to known genes (Table 5). Among the annotated gene list, the ID1 gene highlights as biological candidate to underlay the QTL effects. Studies in mice suggest that ID1 is a negative regulator of insulin secretion, playing an essential role in the etiology of glucose intolerance, insulin secretory dysfunction, and β-cell dedifferentiation under conditions of increased lipid supply [71].
Additionally, we noted that some SNPs within five genes (PTPRD, AGL, VCAM1, VDR, FASL and CYP2E1) considered positional and biological candidates, as it is mentioned in the previous paragraphs, are contained in the PorcineSNP60 BeadChip, according to Sscrofa10.2 annotation. Therefore, these SNPs were tested to underlay the corresponding QTL effects in association analyses (Table 6). The main results of the SNP association were for two of the AGL SNPs with BFT75 in the BC1 generation. Even more, a haplotype analysis of the two AGL SNPs (haplotypes: ASGA0022526G-ALGA0028692C (H1), ASGA0022526A-ALGA0028692G (H2) and ASGA0022526A-ALGA0028692C (H3)) revealed higher significant effects than the single SNP analyses (P-value = 4.6 x 10-6). The H1 haplotype showed the strongest effect (1.02 ± 0.22 mm). These SNPs are located in non-coding regions and they are likely in linkage disequilibrium with the causative mutation underlying the QTL effects in SSC4.
Conclusions
The arrival of the high-density SNP panels makes available high-resolution linkage maps increasing the information content for the successful QTL identification. In the current study, the use of the PorcineSNP60 BeadChip has allowed to detect significant QTL for fatness and yield cuts in ten autosomes (SSC1, SSC2, SSC4, SSC5, SSC6, SSC9, SSC11, SSC13, SSC14 and SSC17). Two of the QTL regions, in SSC4 and SSC6, had been previously identified in the same animal material, however, the remaining ones were not previously detected probably due to the limited number of microsatellite markers employed in those scans. Moreover, most of the significant QTL regions displayed narrow CI making easier the selection of candidate genes. Finally, prominent putative biological and positional candidate genes underlying those QTL effects are listed based on recent porcine genome annotation.
Abbreviations
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
- LEPR:
-
Leptin receptor
- MTTP:
-
Microsomal triglyceride transfer protein
- FABP5:
-
Fatty acid binding protein 5
- TYRP1:
-
Tyrosinase-related protein 1
- PTPRD:
-
Protein tyrosine phosphatase, receptor type, D
- PPARGC1B:
-
Peroxisome proliferator-activated receptor gamma, coactivator 1 beta
- AGL:
-
Amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
- VCAM1:
-
Vascular cell adhesion molecule 1
- ADIPOR2:
-
Adiponectin receptor 2
- VDR:
-
Vitamin D (1,25- dihydroxyvitamin D3) receptor
- AMPK:
-
Protein kinase, AMP-activated, beta 1 non-catalytic subunit
- MYOG:
-
Myogenin (myogenic factor 4)
- FASL:
-
Fas ligand (TNF superfamily, member 6)
- CYP2E1:
-
Cytochrome P450, family 2, subfamily E, polypeptide 1
- NCK1:
-
NCK adaptor protein 1
- BPIF:
-
BPI fold containing family C
- ID1:
-
Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein.
References
Fujii J, Otsu K, Zorzato F, de Leon S, Khanna VK, Weiler JE, O’Brien PJ, MacLennan DH: Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science. 199, 253: 448-451.
Milan D, Jeon JT, Looft C, Amarger V, Robic A, Thelander M, Rogel-Gaillard C, Paul S, Iannuccelli N, Rask L, Ronne H, Lundström K, Reinsch N, Gellin J, Kalm E, Roy PL, Chardon P, Andersson L: A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000, 19: 1248-1251.
Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L: A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003, 425: 832-836. 10.1038/nature02064.
Ciobanu DC, Bastiaansen J, Lonergan SM, Thomsen H, Dekkers JCM, Plastow GS, Rothschild MF: New alleles in calpastatin gene are associated with meat quality traits in pigs J Anim Sci. 2004, 82: 2829-2839.
Andersson L, Georges M: Domestic-animal genomics: deciphering the genetics of complex traits: Nat Rev Genet. 2004, 5: 202-212.
John S, Shephard N, Liu G, Zeggini E, Cao M, Chen W, Vasavda N, Mills T, Barton A, Hinks A, Eyre S, Jones KW, Ollier W, Silman A, Gibson N, Worthington J, Kennedy GC: Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. Am J Hum Genet. 2004, 75: 54-64. 10.1086/422195.
Pérez-Enciso M, Varona L: Quantitative trait loci mapping in F2 crosses between outbred lines. Genetics. 2000, 155: 391-405.
Varona L, Óvilo C, Clop A, Noguera JL, Pérez-Enciso M, Coll A, Folch JM, Barragán C, Toro MA, Babot D, Sánchez A: QTL mapping for growth and carcass traits in an Iberian by Landrace pig intercross: additive, dominant and epistatic effects. Genet Res. 2002, 80: 145-154.
Mercadé A, Estellé J, Noguera JL, Folch JM, Varona L, Silió L, Sánchez A, Pérez-Enciso M: On growth, fatness, and form: a further look at porcine chromosome 4 in an Iberian x Landrace cross. Mamm Genome. 2005, 16: 374-382. 10.1007/s00335-004-2447-4.
Óvilo C, Pérez-Enciso M, Barragan C, Clop A, Rodriguez C, Oliver MA, Toro MA, Noguera JL: A QTL for intramuscular fat and backfat thickness is located on porcine chromosome 6. Mamm Genome. 2000, 11: 344-346. 10.1007/s003350010065.
Óvilo C, Oliver A, Noguera JL, Clop A, Barragán C, Varona L, Rodríguez C, Toro MA, Sánchez A, Pérez-Enciso M, Silió L: Test for positional candidate genes for body composition on pig chromosome 6. Genet Sel Evol. 2002, 34: 465-479. 10.1186/1297-9686-34-4-465.
Óvilo C, Fernández A, Noguera JL, Barragán C, Letón R, Rodríguez C, Mercadé A, Alves E, Folch JM, Varona L, Toro MA: Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet Res. 2005, 85: 57-67. 10.1017/S0016672305007330.
Óvilo C, Fernández A, Fernández AI, Folch JM, Varona L, Benítez R, Nuñez Y, Rodríguez C, Silió L: Hypothalamic expression of porcine leptin receptor (LEPR), neuropeptide Y (NPY), and cocaine- and amphetamine-regulated transcript (CART) genes is influenced by LEPR genotype. Mamm Genome. 2010, 21: 583-591. 10.1007/s00335-010-9307-1.
Estellé J, Fernández AI, Pérez-Enciso M, Fernández A, Rodríguez C, Sánchez A, Noguera JL, Folch JM: A non-synonymous mutation in a conserved site of the MTTP gene is strongly associated with protein activity and fatty acid profile in pigs. Anim Genet. 2009, 40: 813-820. 10.1111/j.1365-2052.2009.01922.x.
Estellé J, Pérez-Enciso M, Mercadé A, Varona L, Alves E, Sánchez A, Folch JM: Characterization of the porcine FABP5 gene and its association with the FAT1 QTL in an Iberian by Landrace cross. Anim Genet. 2006, 37: 589-591. 10.1111/j.1365-2052.2006.01535.x.
Mercadé A, Pérez-Enciso M, Varona L, Alves E, Noguera JL, Sánchez A, Folch JM: Adipocyte fatty-acid binding protein is closely associated to the porcine FAT1 locus on chromosome 4. J Anim Sci. 2006, 84: 2907-2913. 10.2527/jas.2005-663.
Evans DM, Cardon LR: Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet. 2004, 75: 687-692. 10.1086/424696.
Gonzalez-Neira A, Rosa-Rosa JM, Osorio A, Gonzalez E, Southey M, Sinilnikova O, Lynch H, Oldenburg RA, van Asperen CJ, Hoogerbrugge N, Pita G, Devilee P, Goldgar D, Benitez J: Genomewide high-density SNP linkage analysis of non-BRCA1/2 breast cancer families identifies various candidate regions and has greater power than microsatellite studies. BMC Genomics. 2007, 8: 299-10.1186/1471-2164-8-299.
Chioza BA, Aicardi J, Aschauer H, Brouwer O, Callenbach P, Covanis A, Dooley JM, Dulac O, Durner M, Eeg-Olofsson O, Feucht M, Friis ML, Guerrini R, Kjeldsen MJ, Nabbout R, Nashef L, Sander T, Sirén A, Wirrell E, McKeigue P, Robinson R, Gardiner RM, Everett KV: Genome wide high density SNP-based linkage analysis of childhood absence epilepsy identifies a susceptibility locus on chromosome 3p23-p14. Epilepsy Res. 2009, 87: 247-255. 10.1016/j.eplepsyres.2009.09.010.
Ramos AM, Crooijmans RP, Affara NA, Amaral AJ, Archibald AL, Beever JE, Bendixen C, Churcher C, Clark R, Dehais P, Hansen MS, Hedegaard J, Hu ZL, Kerstens HH, Law AS, Megens HJ, Milan D, Nonneman DJ, Rohrer GA, Rothschild MF, Smith TP, Schnabel RD, Van Tassell CP, Taylor JF, Wiedmann RT, Schook LB, Groenen MA: Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009, 4: e6524-10.1371/journal.pone.0006524.
Muñoz M, Alves E, Ramayo-Caldas Y, Casellas J, Rodríguez C, Folch JM, Silió L, Fernández AI: Recombination rates across porcine autosomes inferred from high-density linkage maps. Anim Genet. 2011, 10.1111/j.1365-2052.2011.02301.x.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007, 81: 559-275. 10.1086/519795.
Storey JD, Taylor JE, Siegmund D: Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J R Stat Soc Series B. 2004, 66: 187-205. 10.1111/j.1467-9868.2004.00439.x.
Mangin B, Goffinet B, Rebaï A: Constructing confidence intervals for QTL location. Genetics. 1994, 138: 1301-1308.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21: 263-265. 10.1093/bioinformatics/bth457.
Pérez-Enciso M: Misztal I. Qxpak.5: old mixed model solutions for new genomics problems. BMC Bioinformatics. 2011, 12: 202-
Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, Andersson K, Andersson-Eklund L, Edfors-Lilja I, Fredholm M, Hansson I, et al: Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science. 1994, 263: 1771-1774. 10.1126/science.8134840.
Marklund L, Nyström PE, Stern S, Andersson-Eklund L, Andersson L: Confirmed quantitative trait loci for fatness and growth on pig chromosome 4. Heredity. 1999, 82: 134-141. 10.1038/sj.hdy.6884630.
Fan B, Onteru SK, Du ZQ, Garrick DJ, Stalder KJ, Rothschild MF: Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs. PLoS One. 2011, 6: e14726-10.1371/journal.pone.0014726.
Rohrer GA, Keele JW: Identification of quantitative trait loci affecting carcass composition in swine: I. Fat deposition traits. J Anim Sci. 1998, 76: 2247-2254.
Rohrer GA: Identification of quantitative trait loci affecting birth characters and accumulation of backfat and weight in a Meishan-White Composite resource population. J Anim Sci. 2000, 78: 2547-2553.
Bidanel JP, Milan D, Iannuccelli N, Amigues Y, Boscher MY, Bourgeois F, Caritez JC, Gruand J, Le Roy P, Lagant H, Quintanilla R, Renard C, Gellin J, Ollivier L, Chevalet C: Detection of quantitative trait loci for growth and fatness in pigs. Genet Sel Evol. 2001, 33: 289-309. 10.1186/1297-9686-33-3-289.
Geldermann H, Cepica S, Stratil A, Bartenschlager H, Preuss S: Genome-wide mapping of quantitative trait loci for fatness, fat cell characteristics and fat metabolism in three porcine F2 crosses. Genet Sel Evol. 2010, 42: 31-10.1186/1297-9686-42-31.
Quintanilla R, Milan D, Bidanel JP: A further look at quantitative trait loci affecting growth and fatness in a cross between Meishan and Large White pig populations. Genet Sel Evol. 2002, 34: 193-210. 10.1186/1297-9686-34-2-193.
Knott SA, Marklund L, Haley CS, Andersson K, Davies W, Ellegren H, Fredholm M, Hansson I, Hoyheim B, Lundström K, Moller M, Andersson L: Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics. 1998, 149: 1069-1080.
Sławińska A, Siwek M, Knol EF, Roelofs-Prins DT, van Wijk HJ, Dibbits B, Bednarczyk M: Validation of the QTL on SSC4 for meat and carcass quality traits in a commercial crossbred pig population. J Anim Breed Genet. 2009, 126: 43-51. 10.1111/j.1439-0388.2008.00753.x.
Edwards DB, Ernst CW, Tempelman RJ, Rosa GJ, Raney NE, Hoge MD, Bates RO: Quantitative trait loci mapping in an F2 Duroc x Pietrain resource population: I. Growth traits. J Anim Sci. 2008, 86: 241-253.
Milan D, Bidanel JP, Iannuccelli N, Riquet J, Amigues Y, Gruand J, Le Roy P, Renard C, Chevalet C: Detection of quantitative trait loci for carcass composition traits in pigs. Genet Sel Evol. 2002, 34: 705-728. 10.1186/1297-9686-34-6-705.
Guo T, Ren J, Yang K, Ma J, Zhang Z, Huang L: Quantitative trait loci for fatty acid composition in longissimus dorsi and abdominal fat: results from a White Duroc x Erhualian intercross F2 population. Anim Genet. 2009, 40: 185-191. 10.1111/j.1365-2052.2008.01819.x.
Liu G, Kim JJ, Jonas E, Wimmers K, Ponsuksili S, Murani E, Phatsara C, Tholen E, Juengst H, Tesfaye D, Chen JL, Schellander K: Combined line-cross and half-sib QTL analysis in Duroc-Pietrain population. Mamm Genome. 2008, 19: 429-438. 10.1007/s00335-008-9132-y.
de Koning DJ, Harlizius B, Rattink AP, Groenen MA, Brascamp EW, van Arendonk JA: Detection and characterization of quantitative trait loci for meat quality traits in pigs. J Anim Sci. 2001, 79: 2812-2819.
Xue HL, Zhou ZX: Effects of the MyoG gene on the partial growth traits in pigs. Yi Chuan Xue Bao. 2006, 33: 992-997.
Kim CW, Hong YH, Yun SI, Lee SR, Kim YH, Kim MS, Chung KH, Jung WY, Kwon EJ, Hwang SS, Park DH, Cho KK, Lee JG, Kim BW, Kim JW, Kang YS, Yeo JS, Chang KT: Use of microsatellite markers to detect quantitative trait loci in Yorkshire pigs. J Reprod Dev. 2006, 52: 229-237. 10.1262/jrd.17046.
Liu G, Jennen DG, Tholen E, Juengst H, Kleinwächter T, Hölker M, Tesfaye D, Un G, Schreinemachers HJ, Murani E, Ponsuksili S, Kim JJ, Schellander K, Wimmers K: A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim Genet. 2007, 38: 241-252. 10.1111/j.1365-2052.2007.01592.x.
Xu XL, Xu XW, Pan PW, Li K, Jiang ZH, Yu M, Rothschild MF, Liu B: Porcine skeletal muscle differentially expressed gene CMYA1: isolation, characterization, mapping, expression and association analysis with carcass traits. Anim Genet. 2009, 40: 255-261. 10.1111/j.1365-2052.2008.01825.x.
Ramos AM, Bastiaansen JW, Plastow GS, Rothschild MF: Genes located on a SSC17 meat quality QTL region are associated with growth in outbred pig populations. Anim Genet. 2009, 40: 774-778. 10.1111/j.1365-2052.2009.01907.x.
Pierzchala M, Cieslak D, Reiner G, Bartenschlager H, Moser G, Geldermann H: Linkage and QTL mapping for Sus scrofa chromosome 17. J Anim Breed Genet. 2003, 120: 132-137.
Russo V, Fontanesi L, Scotti E, Beretti F, Davoli R: Nanni Costa L, Virgili R, Buttazzoni L: Single nucleotide polymorphisms in several porcine cathepsin genes are associated with growth, carcass, and production traits in Italian Large White pigs. J Anim Sci. 2008, 86: 3300-3314. 10.2527/jas.2008-0920.
Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, Chang CC, Chen P, Chen CH, Fann CS, Chen YT, Wu JY: A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010, 6: e1000847-10.1371/journal.pgen.1000847.
Handschin C, Spiegelman BM: Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006, 27: 728-735.
Silva KM, Bastiaansen JW, Knol EF, Merks JW, Lopes PS, Guimarães SE, van Arendonk JA: Meta-analysis of results from quantitative trait loci mapping studies on pig chromosome 4. Anim Genet. 2011, 42: 280-292. 10.1111/j.1365-2052.2010.02145.x.
Fontanesi L, Scotti E, Buttazzoni L: Dall'Olio S, Davoli R, Russo V: A single nucleotide polymorphism in the porcine cathepsin K (CTSK) gene is associated with back fat thickness and production traits in Italian Duroc pigs. Mol Biol Rep. 2010, 37: 491-495. 10.1007/s11033-009-9678-0.
Ojeda A, Estellé J, Folch JM, Pérez-Enciso M: Nucleotide variability and linkage disequilibrium patterns at the porcine FABP5 gene. Anim Genet. 2008, 39: 468-473. 10.1111/j.1365-2052.2008.01752.x.
Han SH, Shin KY, Lee SS, Ko MS, Jeong DK, Oh HS, Yang BC, Cho IC: SINE indel polymorphism of AGL gene and association with growth and carcass traits in Landrace x Jeju Black pig F(2) population. Mol Biol Rep. 2010, 37: 467-471. 10.1007/s11033-009-9644-x.
Fontanesi L, Galimberti G, Calò DG, Fronza R, Martelli PL, Scotti E, Colombo M, Schiavo G, Casadio R, Buttazzoni L, Russo V: Identification and association analysis of several hundred single nucleotide polymorphisms within candidate genes for backfat thickness in Italian Large White pigs using a selective genotyping approach. J Anim Sci. 2012, : -[Epub ahead of print]
Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, Sugiyama T, Miyagishi M, Hara K, Tsunoda M, Murakami K, Ohteki T, Uchida S, Takekawa S, Waki H, Tsuno NH, Shibata Y, Terauchi Y, Froguel P, Tobe K, Koyasu S, Taira K, Kitamura T, Shimizu T, Nagai R, Kadowaki T: Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003, 423: 762-769. 10.1038/nature01705.
Grundberg E, Brändström H, Ribom EL, Ljunggren O, Mallmin H, Kindmark A: Genetic variation in the human vitamin D receptor is associated with muscle strength, fat mass and body weight in Swedish women. Eur J Endocrinol. 2004, 150: 323-328. 10.1530/eje.0.1500323.
Lopez-Buesa P, Óvilo C, Rodríguez MC, Varona L, Silió L, Burgos C, Galve A: The effects of leptin receptor (LEPR) and melanocortin-4 receptor (MC4R) polymorphisms in fat content, fat distribution, and fat composition in a Duroc x Landrace/Large White cross. Livestock Science. 2011, 10.1016/j.livsci.2012.01.010.
Muñoz G, Óvilo C, Silió L, Tomás A, Noguera JL, Rodriguez MC: Single- and joint population analyses of two experimental pig crosses to confirm quantitative trait loci on Sus scrofa chromosome 6 and leptin receptor effects on fatness and growth traits. J Anim Sci. 2009, 87: 459-468.
Muñoz G, Alcázar E, Fernández A, Barragán C, Carrasco A, de Pedro E, Silió L, Sánchez JL, Rodríguez MC: Effects of porcine MC4R and LEPR polymorphisms, gender and Duroc sire line on economic traits in Duroc x Iberian crossbred pigs. Meat Sci. 2011, 88: 169-173. 10.1016/j.meatsci.2010.12.018.
Rodriguez MC, Fernandez A, Carrasco C, Garcia A, Gomez E, De Mercado E, Lopez MA, Óvilo C, Silio L: Effect of LEPR c.2002 C>T SNP on feed intake and growth in heavy Duroc X Iberian crossbred pigs. In Proceedings of the ninth world congress on genetics applied to livestock production. 2010, : 1-6. Leipzig
Krzęcio E, Koćwin-Podsiadła M, Kurył J, Zybert A, Sieczkowska H, Antosik K: The effect of interaction between genotype CAST/RsaI (calpastatin) and MYOG/MspI (myogenin) on carcass and meat quality in pigs free of RYR1(T) allele. Meat Sci. 2008, 80: 1106-1115. 10.1016/j.meatsci.2008.05.002.
Wyszyńska-Koko J, Pierzchała M, Flisikowski K, Kamyczek M, Rózycki M, Kurył J: Polymorphisms in coding and regulatory regions of the porcine MYF6 and MYOG genes and expression of the MYF6 gene in m. longissimus dorsi versus productive traits in pigs. J Appl Genet. 2006, 47: 131-138. 10.1007/BF03194612.
Ikeda T, Kanazawa T, Otsuka S, Ichii O, Hashimoto Y, Kon Y: Expression of caspase family and muscle- and apoptosis-specific genes during skeletal myogenesis in mouse embryo. J Vet Med Sci. 2009, 71: 1161-1168. 10.1292/jvms.71.1161.
Squires EJ: Lundström: Relationship between cytochrome P450IIE1 in liver and levels of skatole and its metabolites in intact male pigs. J Anim Sci. 1997, 75: 2506-2511.
Babol J, Squires EJ, Lundström K: Hepatic metabolism of skatole in pigs by cytochrome P4502E1. J Anim Sci. 1998, 76: 822-828.
Lin Z, Lou Y, Squires EJ: Functional polymorphism in porcine CYP2E1 gene: Its association with skatole levels. J Steroid Biochem Mol Biol. 2006, 99: 231-237. 10.1016/j.jsbmb.2005.07.001.
Zong H, Armoni M, Harel C, Karnieli E, Pessin JE: Cytochrome P450 CYP2E1 knockout mice are protected against high fat diet induced obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2011, 10.1152/ajpendo.00258.2011.
Aubert J, Begriche K, Knockaert L, Robin MA, Fromenty B: Increased expression of cytochrome P450 2E1 in nonalcoholic fatty liver disease: mechanisms and pathophysiological role. Clin Res Hepatol Gastroenterol. 2011, 35: 630-637. 10.1016/j.clinre.2011.04.015.
Latreille M, Laberge MK, Bourret G, Yamani L, Larose L: Deletion of Nck1 attenuates hepatic ER stress signaling and improves glucose tolerance and insulin signaling in liver of obese mice. Am J Physiol Endocrinol Metab. 2011, 300: E423-434. 10.1152/ajpendo.00088.2010.
Akerfeldt MC, Laybutt DR: Inhibition of Id1 augments insulin secretion and protects against high-fat diet-induced glucose intolerance. Diabetes. 2011, 60: 2506-2514. 10.2337/db11-0083.
Acknowledgements
This work was funded by MICINN projects AGL2008-04818-C03/GAN and CSD2007-00036. DPM was funded by a FPI Ph.D grant from the Spanish Ministerio de Educación (BES-2009-025417). YR was funded by a FPU Ph.D grant from the Spanish Ministerio de Educación (AP2008-01450). We want to thanks to Dr. Martien Groenen (Wageningen, NL) for the SNP annotation on porcine genome assembly, to Anna Mercadé for her technical assistance with the SNPs genotyping and to Rita Benítez and Fabián García for technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AIF, JMF, JLN, MCR and LS conceived and designed the experiment. MCR and AIF performed the data analysis and drafted the manuscript. JLN, NIE, AIF, MCR, LS and JMF collected the samples and records. AC performed the SNP genotyping, CB, AIF and YR performed the SNP filtering. DPM and AIF performed the SNP probe and gene annotation. All authors read and approved the final manuscript.
Electronic supplementary material
12863_2012_1007_MOESM1_ESM.docx
Additional file 1: Table S1. Positions and additive effects of significant QTL at the chromosome-wide level (q-value < 0.10). (DOCX 16 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Fernández, A.I., Pérez-Montarelo, D., Barragán, C. et al. Genome-wide linkage analysis of QTL for growth and body composition employing the PorcineSNP60 BeadChip. BMC Genet 13, 41 (2012). https://doi.org/10.1186/1471-2156-13-41
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-13-41