Abstract
Background
Low nucleotide divergence between human and chimpanzee does not sufficiently explain the species-specific morphological, physiological and behavioral traits. As gene duplication is a major prerequisite for the emergence of new genes and novel biological processes, comparative studies of human and chimpanzee duplicated genes may assist in understanding the mechanisms behind primate evolution. We addressed the divergence between human and chimpanzee duplicated genomic regions by using Luteinizing Hormone Beta (LHB)/Chorionic Gonadotropin Beta (CGB) gene cluster as a model. The placental CGB genes that are essential for implantation have evolved from an ancestral pituitary LHB gene by duplications in the primate lineage.
Results
We shotgun sequenced and compared the human (45,165 bp) and chimpanzee (39,876 bp) LHB/CGB regions and hereby present evidence for structural variation resulting in discordant number of CGB genes (6 in human, 5 in chimpanzee). The scenario of species-specific parallel duplications was supported (i) as the most parsimonious solution requiring the least rearrangement events to explain the interspecies structural differences; (ii) by the phylogenetic trees constructed with fragments of intergenic regions; (iii) by the sequence similarity calculations. Across the orthologous regions of LHB/CGB cluster, substitutions and indels contributed approximately equally to the interspecies divergence and the distribution of nucleotide identity was correlated with the regional repeat content. Intraspecies gene conversion may have shaped the LHB/CGB gene cluster. The substitution divergence (1.8–2.59%) exceeded two-three fold the estimates for single-copy loci and the fraction of transversional mutations was increased compared to the unique sequences (43% versus ~30%). Despite the high sequence identity among LHB/CGB genes, there are signs of functional differentiation among the gene copies. Estimates for dn/ds rate ratio suggested a purifying selection on LHB and CGB8, and a positive evolution of CGB1.
Conclusion
If generalized, our data suggests that in addition to species-specific deletions and duplications, parallel duplication events may have contributed to genetic differences separating humans from their closest relatives. Compared to unique genomic segments, duplicated regions are characterized by high divergence promoted by intraspecies gene conversion and species-specific chromosomal rearrangements, including the alterations in gene copy number.
Similar content being viewed by others
Background
Gene duplication has long been considered as one of the main mechanisms of the adaptive evolution and as an important source of the genetic novelty [1]. Differential duplications and deletions of chromosomal regions including coding genes provide a powerful source for the evolution of species-specific biological differences [2]. Compared to other mammals, the genomes of primates show an enrichment of large segmental duplications with high levels (>90%) of sequence identity [3]. In the human genome particularly pronounced expansions of the copy number have been reported for genes involved in the structure and function of the brain [4]. In comparison of human and its closest relative chimpanzee, large duplications contribute considerably (2.7%; [5]) to the overall divergence compared to single base pair substitutions (1.2–1.5%; [2, 6–12]). In addition to providing the substrate for non-allelic homologous recombination mediating genomic disorders (reviewed by [13]), the duplication architecture of a genome may also influence normal phenotypic variation. It has been estimated that ~20% of segmental duplications are polymorphic within human and chimpanzee populations contributing to intraspecies diversity [5, 14]. Despite the fact that segmental duplications cover a substantial fraction of the great apes genomes, the experimental data on the divergence and detailed evolutionary dynamics of duplicated gene regions is still limited. Sequence comparison of duplicated genes in sister-species would assist in understanding the mechanisms behind primate evolution and in associating the genetic divergence with phenotypic diversification.
One of the genomic regions that has evolved through several gene duplication events in primate lineage is the Luteinizing Hormone Beta (LHB)/Chorionic Gonadotropin Beta (CGB) gene cluster locating in human at 19q13.32. The LHB/CGB genes have an essential role in reproduction: placentally expressed HCG hormone contributes to the implantation process of the embryo during the early stages of pregnancy, pituitary expressed luteinizing hormone promotes the ovulation and luteinization of follicles and stimulates the steroidogenesis. In human, the cluster consists of seven highly homologous genes: an ancestral LHB and six duplicated CGB genes [15]. The data from other primates supports the hypothesis of several sequential duplication events increasing gradually the number of CGB genes among primates from one (New World monkeys: the owl monkey, Aotus trivirgatus and the dusky titi monkey, Callicebus moloch) to six in human (Figure 1A). The mapped copy number of the CGB gene among Old World monkeys varies: three in rhesus macaque (Macaca mulatta), five in guereza monkey (Colobus guereza) and dusky leaf monkey (Presbytis obscura), four in orangutan (Pongo pygmaeus) [16]. It has been suggested that CGB gene first arose in the common ancestor of the anthropoid primates after diverging from tarsiers [16].
We have chosen LHB/CGB genomic region as a model to study the evolution of recent primate duplications. Although the chimpanzee genome has been sequenced, there are still large gaps and uncertainties concerning the segmental duplications regions, including the LHB/CGB region. To obtain a high-quality DNA sequence, we constructed and sequenced a shot-gun library, and hereby report the complete sequence of the entire LHB/CGB cluster in the common chimpanzee. We are addressing the following aspects regarding to the evolution of duplicated genes in closely related species: (i) in-depth comparison of the human and chimpanzee LHB/CGB genome clusters; (ii) variation in substitution rates; (iii) genic and intergenic divergences; (iv) impact of intraspecies gene conversion in phylogeny, divergence and transversion/transition ratios; (v) evidence of natural selection. To our knowledge, this is the first detailed report of parallel independent duplication events initiated within a duplicated genome cluster and leading to structural divergence in the two sister-species, human and common chimpanzee.
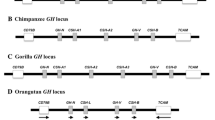
Evolution of the Gonadotropin Hormone Beta ( CGB ) genes. Duplication of ancestral Luteinizing Hormone Beta (LHB) gene in primate lineage has given rise to a novel gene, CGB. (A) A simplified schematic presentation of the evolution of LHB/CGB genes in primates [15, 16]. (B) Comparative structure of the human (GenBank reference: NG_000019) and the chimpanzee LHB/CGB cluster (this study, Genbank: accession number EU000308) drawn to an approximate scale. Coding genes are depicted as wide empty arrows in the direction of transcription on the sense strand. A – E indicate the intergenic regions. B' and C' denote putative duplications of the intergenic regions B and C. Identical color and pattern codes refer to the DNA segments within the cluster with highly similar sequences, the direction of the DNA sequence is indicated on the sense strand. Sequence identity within the cluster: between coding genes 85–99%; intergenic regions A and E 81%; C and C' 96%; B, B' and D ranging 81–98%.
Results and Discussion
Human and chimpanzee LHB/CGBgenome clusters differ considerably in size
The total length of the sequenced chimpanzee (Ch) LHB/CGB genomic region obtained from two overlapping BAC clones was 43,945 bp. It encompasses 1,029 bp of the flanking (centromeric side) RUVBL2 gene, 39,876 bp of the entire LHB/CGB cluster and 2,986 bp of the flanking (telomeric side) NTF5 gene (Figure 1B; Genbank submission: EU000308). Compared to the human (Hu) LHB/CGB region (Genbank: NG_000019), the sequence of the ChLHB/CGB cluster is 5,289 bp shorter. The sequence characteristics of the LHB/CGB genomic regions are similar in these two species: extremely high GC-nucleotide content (57% compared to average 41% for Hu and Ch [10]), high fraction of CpG islands (Hu 6.6%, Ch 6.1% compared to estimated 1–3.5% for Hu and Ch [6, 8]) and repetitive sequences (Hu 26.9%, Ch 25.15%), especially SINEs (Hu 23.23%, Ch 21.81%) (Additional file 1). High repeat content is also characteristic to several other duplicated regions, such as MHC class I region [7] and Apolipoprotein CI genomic segment [17].
As expected, there is a considerable similarity between the genomic organization of human and chimpanzee LHB/CGB clusters (Figure 1B). We identified two highly identical, apparently orthologous segments within the cluster: RUVBL2/LHB/intergenic region A (Ch 8,084 bp, Hu 7,973 bp; 96% sequence identity) and the region spanning from CGB1 to NTF5 (Ch 29,136 bp, Hu 28,568 bp; 94.8% sequence identity). However, a large species-specific structural rearrangement was localized between the intergenic region A and CGB1 gene, resulting in discordant size of human (45,165 bp) and chimpanzee (39,876 bp) clusters as well as species-specific number of duplicated gene copies, seven for human (1 LHB + 6 CGB genes) and six (1 LHB + 5 CGB) for chimpanzee. In human the rearranged region (12,700 bp) harbors one HCG beta coding CGB gene and one CGB1/2-like gene (CGB2) recognized by a specific promoter-segment [18, 19], while in chimp (rearranged region 6,725 bp) only a CGB1/2-like gene (CGB1B) is present in an inverted orientation compared to human. In addition, ChLHB/CGB cluster lacks the whole intergenic region C' and has a considerably shorter inverted intergenic region B' (Figure 1B).
Evidence for independent duplication events within LHB/CGBgene cluster for two closely related primate species
We considered alternative scenarios that may have led to structural differences in LHB/CGB clusters in two sister-species. The scenario of species-specific parallel duplications was supported by several lines of evidence (Figure 1B, Figure 2). First, it was the most parsimonious solution requiring the smallest number of rearrangement events. Assuming that the ancestral Hu-ChLHB/CGB cluster consisted of the present-day highly identical segments (Figure 1B; see above), only one evolutionary event would explain the current structure of ChLHB/CGB cluster – a direct duplication of CGB1 and most of intergenic region B (excluding psNTF6G') giving arise to the segment CGB1B/Ch-B'. In humans, two events would have lead to present HuLHB/CGB (not in order) – an inverted duplication of the entire region from CGB1 to CGB5 gene (segment from CGB to CGB2) and a direct duplication and translocation of region C creating C' next to CGB1 gene (Figure 1B). These parallel duplications in two sister-species might have been initiated by non-allelic homologous recombinations between multiple Alu SINE sequences (AluSx, AluSp, AluSq) locating at the junctions of both human- and chimp-specific duplications. Consistently, the phylogenetic trees that were constructed using different fragments of intergenic regions show that Ch-B and Ch-B', Hu-B and Hu-B' as well as Hu-C and Hu-C' clearly cluster together supporting their paralogous status (Figure 2B–F). The Hu-D and Ch-D region form a well-supported clade (Figure 2B–D, G) giving evidence that these are orthologous segments. The phylogenetic relationship supports the ancestral status of region C (present in both species) compared to Hu-C' (human-specific duplication). Also the sequence similarity calculations are consistent with the scenario of independent species-specific duplications: the identity of Ch-B and Ch-B' regions is 98.1%, and of ChCGB1 and ChCGB1B 98.7%. The latter exceeds the estimates for any orthologous genes between human and chimp in LHB/CGB cluster (from 98.2% in LHB to 97.4% in CGB5 and CGB8). However, the possibility of more intensive gene conversion between these intergenic segments resulting in higher intraspecies similarity cannot be excluded. It is well known that past intraspecies gene conversion events might be reflected by tree phylogenies and could lead to erroneous conclusions [16, 20]. The footprint of gene conversion is also reflected on the phylogenic tree of human and chimpanzee LHB/CGB genes (Figure 2A). Instead of two separate clades for orthologous CGB5 and CGB8, the genes within one species cluster together.
Neighbor-joining trees based on genic (A) and intergenic (B-G) regions within LHB/CGB gene cluster. (A) A phylogenetic tree of the full sequences of LHB/CGB gene from Homo sapiens, Pan troglodytes, Gorilla gorilla and Pongo pygmeaus. (*) denotes sequences from [44]. Phylogenetic analysis of intergenic regions was conducted with segments without (B-D) and with (E-G) covering NTF6 pseudogenes. The homologous segments used for each respective phylogenetic analysis are indicated with a circle on a consensus structure of the intergenic regions in LHB/CGB cluster (boxed; from Figure 1B). The nomenclature of the intergenic regions is as on Figure 1B. Bootstrap support values are shown at the nodes (1000 bootstrap replications). Abbreviations: hu – human, ch – chimpanzee, gor – gorilla, orang – orangutan.
Alternative scenarios leading to discordant gene number in LHB/CGB gene clusters in these two species are less supported. A minimum of three rearrangement events (Figure 1B) would have been required for the chimpanzee-specific deletion: loss of CGB and psNTF6A gene accompanied with the inversion of CGB2 and intergenic region B' (giving rise to CGB1B and chimp B'), and a separate deletion of region C' in chimp. Also the scenario of human-specific duplication (Figure 1B) would have required at least three events: an inversion of CGB1B and intergenic region Ch-B' (giving rise to part of Hu-B' and CGB2); either a direct duplication and translocation of CGB8 gene along with psNTF6A' or an inverted duplication and translocation of CGB5 along with psNTF6G' (resulting in CGB and psNTF6A), and a direct duplication and translocation of intergenic region C creating Hu-C' next to CGB1 gene.
A number of gene families have been characterized where the gene number differs between human and chimpanzee due to species-specific indels [7, 21]. To our knowledge, this is the first report where parallel independent duplications arisen within the same region in human and chimpanzee genomes give the best explanation for the observed structural differences between two sister-species. However, there are examples of independent duplications among primates resulting in convergent functions. A more recent duplication of X-linked opsin gene in New World howler monkeys (Alouatta seniculus and Alouatta caraya) compared to Old World primates, has lead to full trichromacy [22–24] and also there are independently arisen functionally close genes within the Growth Hormone/Somatomammotropin genome cluster in New World monkeys and Old World monkeys/hominoid lineages [20, 25].
Comparative nucleotide divergence profiling of the orthologous regions reveals non-uniform substitution rates across the LHB/CGBregion
We generated a comparative nucleotide divergence profile of substitutions and indels across the orthologous genomic regions of LHB/CGB cluster (Figure 1B, Figure 3). Human (36,541 bp) and chimpanzee (37,220 bp) aligned genomic sequences were analyzed using a non-overlapping sliding window of 500 bp. Several studies have suggested that the majority of the genomic divergence between human and chimpanzee comprises of indels (3.0–11.9%) compared to contribution of nucleotide substitutions (1.2–1.5%) [2, 6–11, 26]. In the duplicated LHB/CGB region indels (2.7%) and substitutions (2.3%) contributed approximately equally to the total divergence (5%) between the two species. In total, 61 indels (mean 16; range 1–637 bp) were identified, 26 as human and 35 as chimpanzee deletions. The size distribution of these indels was consistent with previous reports [2, 6, 9–11] revealing an excess of short indels: 44% involved a single basepair, 77% 1–5 bp and 96.7% <100 bp (Additional files 1 and 2). Two large indels of >100 bp co-locate with repetitive elements: the 128 bp long indel in intergenic region A is located in a simple-repeat rich region; the 637 bp indel in intergenic region B is flanked and composed of SINE sequences (Figure 1B, Figure 3). The latter indel could be defined as a recent human-specific sequence loss since the duplicated human-specific intergenic region B' lacks this deletion. High proportion (>50%) of indels identified between human and chimpanzee has been shown to contain repetitive elements [6, 7]. The regional content of repeats was also correlated with the non-uniform distribution of the nucleotide identity across the cluster (Figure 3; Additional file 1) consistent with the data that repetitive sequences, e.g. Alu repeats have a higher rate of base substitutions [27].
Divergence profile between orthologous regions of the human and the chimpanzee LHB/CGB clusters. In total the compared region covered 37,220 bp (Ch)/36,541 (Hu), including 8,084 bp (Ch)/7,973 bp (Hu) from RUVBL2 gene to the end of intergenic region A and 29,136 bp (Ch)/28,568 bp (Hu) from CGB1 to NTF5 gene. The species-specific large duplications (human 12,700 bp, chimp 6,725 bp) have been excluded from the comparison. The percents of nucleotide substitutions and indels are calculated per 500 bp non-overlapping windows. Grey arrows indicate the locations of coding genes drawn to an approximate scale. A – E denote intergenic regions from Figure 1B. Intergenic repeat fraction includes SINEs, LINEs, satellites, simple repeats and low complexity DNA sequences within each intergenic region.
Transition to transversion ratio in duplicated regions differs from the estimations for unique genomic sequences: possible role of gene conversion
We investigated the distribution of nucleotide substitutions within orthologous regions of LHB/CGB cluster in more detail (Figure 4). Transitions (C⇔T, A⇔G) and transversions (T, C⇔A, G) were found to contribute 62% and 38% of the total substitutions in five orthologous LHB/CGB genes, respectively (Figure 4A). The corresponding estimates for the whole orthologous region (including genes) were 57% for transitions and 43% for transversions (Figure 4B). Notably, the contribution of transitions is ~10% lower than reported in previous studies comparing human and chimpanzee genomic regions (68.87% – 70.3%) [7, 8]. The most frequent transversions are G⇔C substitutions, contributing 16% of all substitutions in genes and 18% across entire orthologous region, exceeding previous estimations by twofold (9.14%, 9%) [7, 8]. Consistently, a large excess of G⇔C transversional pairs as compared to other substitutions has been reported for human HSP70 and mouse Hsp70 orthologous duplicate genes [28]. The high proportion of transversions in the LHB/CGB cluster can be explained by biased gene conversion [29] that leads to a high GC content (57% in human and chimp LHB/CGB region) and thus increases the probability of G⇔C substitutions due to the altered base composition [28, 30, 31].
Profile of nucleotide substitutions in human and chimpanzee orthologous LHB/CGB genes. Grouping of nucleotide substitutions: (A) Nucleotide substitutions in orthologous LHB, CGB1, CGB5, CGB8 and CGB7 genes (in total 6,878 bp; GC-nucleotide content 64%; 161 substitutions). (B) Nucleotide substitutions in the whole orthologous region of the LHB/CGB genome cluster (in total 36,211 bp, GC-nucleotide content 57%, 835 substitutions). Percents for all substitution types are shown with summarized information for transversions and transitions.
Among the transitions, we observed an excess of C⇔T substitutions in LHB/CGB genes (38%) versus the whole region (28%). It is generally accepted that a high proportion of transitions are C to T substitutions in CpG dinucleotides, exhibiting about 10 times higher mutation rate than the genomic average [9, 32]. A higher GC content (64% vs 57%) and presence of CpG islands could explain an excess of C⇔T substitutions in LHB/CGB genes compared to intergenic regions.
Sequence divergence between human and chimpanzee duplicated LHB/CGBgenes is higher than estimates for single copy genes
We studied the human and the chimpanzee orthologous genes LHB, CGB1, CGB5, CGB8 and CGB7 for nucleotide divergence. The nucleotide divergence ranged from 1.8% for LHB to 2.59% for CGB5 and CGB8 genomic sequences (Figure 5; Additional file 3). Despite that the coding regions are most conserved among the species, the exonic divergence rates (mean 1.39%; range 1 – 1.88%) exceeded many times the previous estimates for single-copy regions [8–10, 12, 26, 33]. In non-coding regions, the average nucleotide divergence for promoter regions was as high as 3.22% (range 1 – 5.1%), for introns 2.62% (range 2.04 – 3.24%) and for 5' UTR 2.54% (range 0 – 3.83%). The comparative estimates for single-copy genes are much lower: in promoters 0.75–0.88%, in exons 0.51–1.09%, in introns 1.03–1.47% and in 5' UTR 1.00–1.41% [10–12, 26, 33–36]. It has been suggested that interaction of selection and gene conversion contributes to a higher divergence and diversity in multigene families compared to single-copy genes [37, 38]. As any de novo mutation has a potential to be spread by gene conversion from the original locus to other gene copies, every duplicate could accumulate substitutions arisen in neighboring genes.
Nucleotide divergence (%) between human and chimpanzee orthologous LHB and CGB genes calculated for promoter, 5'UTR, genic, mRNA, exonic and intronic regions. Divergence was estimated by using the human (GenBank: NG_000019) and chimpanzee (this study, Genbank: EU000308) reference sequences alone or by incorporating the diversity data obtained from re-sequencing for one or both species into the calculations. The re-sequencing data for human (n = 95) originated from the published study[29] and for chimpanzee (n = 11) from the unpublished dataset of the authors.
Notably, when the polymorphism data from re-sequencing studies of the human (n = 95 individuals) [29] and the chimpanzee (n = 11 individuals) [Hallast et al, unpublished] LHB/CGB genes were incorporated into calculations, the sequence divergence in genes dropped from 1.8% to 1.26% in LHB, from 2.59% to 1.84% in CGB5, from 2.59% to 1.9% in CGB8 and from 2.18% to 1.57% in CGB7 (Figure 5; Additional file 3). Still, a higher divergence compared to the published data for single-copy genes remained.
Most importantly, our data indicates that the divergence estimates between human and chimpanzee might be substantially lower than reported when the intraspecies variation is taken into account.
Duplicated, highly homologous LHB/CGBgenes evolve under different selective constraints
A number of non-synonymous changes were identified in human-chimpanzee comparison: two in LHB, two in CGB8, three in CGB5 and CGB7 and four in CGB1 (Additional file 3). None of the differences found in exonic regions in chimp altered the ORF nor created a preliminary stop-codon compared to human genes. The maximum likelihood (ML) method was used to estimate ω, non-synonymous (dn)/synonymous (ds) rate ratio for the orthologous genes (n = 5) in the human and chimpanzee LHB/CGB gene cluster (Table 1) [39]. For four of the compared genes (LHB, CGB5, CGB8, CGB7) the parameter was ω < 1, indicating purifying selection. An especially low ω estimate was obtained for LHB (ω = 0.087; 6 synonymous, 2 non-synonymous changes; amino acid divergence 1.42%) and CGB8 (ω = 0.099; 5 synonymous, 2 non-synonymous changes; amino acid divergence 1.21%). Consistently, the dn/dsratio calculated by an alternative Li93 method [40, 41] was statistically significant (dn/ds = 0.146; p = 0.049 two-tailed Z-test [42]) for LHB and reached borderline significance for the CGB8 (dn/ds = 0.162; p = 0.078) (Additional file 4). Indeed, the LHB gene is highly conserved throughout vertebrate evolution in association with its function – it is coding for the receptor-binding beta subunit of luteinizing hormone (LH) critical for successful reproduction [15]. The functional constraint of the human CGB8 may be associated with its major role in contributing to the HCG beta subunit mRNA transcript pool compared to other CGB genes [43].
CGB1 gene (ω = 2.658, 1 synonymous, 4 non-synonymous changes; amino acid divergence 3.03%) stood out as the only locus in the gene cluster with estimated ω >1, which would be consistent with positive or adaptive evolution. It has been suggested that CGB1 has arisen in the common ancestor of African great apes through a duplication event accompanied by an insertion of a novel putative promoter, 5'UTR and exon 1. So far, the detection of CGB1 gene has been unsuccessful in orangutan Pongo pygmaeus by using the PCR approach [44] and in macaque Macaca mulatta (Genbank: AC202849) by in silico search of the current genome assembly. It has been shown that in human the contributions of CGB1 and its duplicate human-specific CGB2 to the summarized expression of the six CGB genes in placenta is much lower (1/1000 to 1/10000) compared to their gene dosage (two genes out of six total) [43, 45]. However, in testis the proportional contribution of CGB1/2 to the total CGB transcript pool is as high as 1/3 [45], which may indicate a possible role of these genes in male reproductive tract. Indeed, a recent study has shown that HCG alpha and HCG beta free subunits are produced in high amounts in the prostate and testes and are subsequently observed in seminal plasma [46].
In order to address which parts of the studied genes exhibit signals of evolving under positive selection, we used CRANN analysis calculating dn and ds values for sliding and overlapping windows along individual genes (Additional file 5) [47, 48]. In case of HCG beta subunit coding genes (CGB5, CGB8 and CGB7), the patterns of nucleotide differences between the sister species were similar – across the protein the synonymous substitutions exceeded the non-synonymous ones and were concentrated in the N- and C-terminus of the protein. In contrast, in CGB1 the non-synonymous substitutions dominated and were distributed in the signal peptide (amino acids 1–20) and the centre of the protein.
Impact of gene duplications in primate divergence
Primate-specific gene duplications have involved loci regulating immunity (e.g. MHC, beta-defensin, CD33rSiglec gene clusters), reproduction (e.g. LHB/CGB, GH/CSH, PRAME genes; Y-chromosomal gene families), development and adaptation (e.g. Beta Globin, Opsin, Rh blood group, Class 1 ADH, PRDM and FAM90A gene families), and brain functions (e.g. NAIP, ROCK1, USP10 and MGC8902 genes) [4, 16, 20, 22, 49–59]. It has been suggested that these duplication events may have been facilitated by non-allelic homologous recombination between Alu sequences, expanded into millions of copies all over primate genomes [54, 60]. There are several examples of independent duplication events in the New World monkey (NWM) and Old World monkey (OWM)/hominoid lineages as well as in distinct primate species. For example, the OWM and apes have three Opsin genes and are trichromats due to gene duplication at the base of the OWM lineage. In NWM, the situation is more variable: most species exhibit two Opsin genes, but in the howler monkey an additional gene duplication has led to full trichromacy [23, 24]. In Growth Hormone gene cluster (five to eight gene copies) some of the duplicate genes in the OWM/hominoid lineages have acquired a novel function and code for Chorionic Somatomammotropin (CSH genes) involved in the glucose metabolism of the fetus and the mother. However, the CSH genes are missing from the genomes of NW monkeys [61]. Further species-specific duplications of GH/CSH genes have been reported for gibbon, macaque, chimpanzee and human [62, 63]. In human-chimpanzee comparison, only three GH/CSH genes are clearly orthologous [21]. Other gene clusters with independent gene duplications in human, apes and macaque lineages are MHC and testes-expressed PRAME genes [53, 64]. The ancestral MHC-B duplicated into MHC-B and MHC-C in hominoids. While human MHC-C orthologs are found in African apes and orangutans, they are not present in macaque or any other OW monkeys [65, 66]. Species-specific evolutionary scenarios have also been reported for LHB/CGB genes [16]. In addition to the structural differences between human and chimpanzee LHB/CGB genes reported in this study, an expansion of CGB genes up to 50 gene copies has been shown in gorilla (Gorilla gorilla) [4, 49]. Despite the high number of structural differences that has been shown between human and chimpanzee genomes by in silico whole-genome analysis (reviewed in [67]), the experimental data for copy number differences between these sister-species has been reported only for a few gene clusters (e.g. GH/CSH, LHB/CGB, PRAME, MGC8902, CXYorf1, KGF, CD33rSiglec, NANOG) [16, 20, 52, 53, 59, 68–70].
In addition to creating structural divergence among the species, duplications provide also the bases for diversification of gene functions. For primate-specific gene duplications, there is evidence of variability in evolutionary rates among the gene copies within and among the species, and of different selective constraints acting on different members of the gene clusters, such as MHC, beta-globin, GH/CSH, PRAME, Rh blood group, CD33rSiglec and beta-defensin genes [71–73]. For example, in human-chimpanzee comparison, the chimp MHC class I loci A, B and C are characterized by lower intra-species allelic variation compared to human, providing evidence that ancestral chimpanzee populations may have experienced a selective sweep [74, 75]. In marmoset LHB/CGB region, a switch of functions has happened between the ancestral LHB and the derived CGB gene. Although LHB and CGB genes are both present at the genomic level, only CGB gene is expressed in the pituitary and placenta tissues. Thus, Chorionic Gonadotropin Hormone is the only gonadotropin carrying also the luteinizing function fulfilled by Luteinizing Hormone in mammals [76, 77].
Duplicated genes tend to evolve in consort facilitated by active inter-locus gene conversion increasing and preserving sequence similarity among the gene copies [37, 38, 78]. Concerted evolution within species may lead to erroneous phylogenetic trees and to the overestimation of interspecies divergence dates [20, 79, 80]. Incorporation of gene conversion data into the equations for calculating interspecies divergence may be still a challenge requiring detailed knowledge of a particular genomic region.
So far, only a limited number of reports has been published that focus on detailed variation patterns of duplicated gene families in primates (MHC, Globin, GH/CSH and LHB/CGB genes) [7, 29, 81–83]. However, the common observation is that compared to single-copy segments, duplicated regions tend to exhibit higher interspecies diversity that could be explained by relaxed selective pressures and/or gene conversions spreading mutations. Thus, when calculating the divergence in duplicated primate-specific regions, the inclusion or exclusion of intraspecies variation data into equations may have a substantial impact on the divergence estimates.
Conclusion
We compared the human and chimpanzee duplicated LHB/CGB genome clusters and hereby present the detailed evidence for parallel independent duplication events in the two sister-species resulting in discordant number of CGB genes (6 in human, 5 in chimpanzee). To our knowledge, this is the first detailed report of parallel duplications in these sister-species leading to structural divergence. The evolutionary fate of duplicated genes is shaped by the interaction of gene conversion and selection. In LHB/CGB gene cluster, active gene conversion may have contributed to higher interspecies sequence divergence (both genic and intergenic sequences) and altered transition/transversion ratio compared to the single-copy loci. This higher divergence remained when intraspecies variation was taken into account. However, the drop in divergence estimates after incorporating the intraspecies variation data promotes to reanalyze previously studied loci, where the human-chimpanzee divergence may be substantially lower than initially calculated. Despite the high sequence homology among LHB/CGB genes (85–99%), there are signs of functional differentiation among the gene copies. To reconstruct the full evolutionary history of LHB/CGB gene cluster, further studies are required comprising high-quality sequence data from several primate species.
Methods
Bacterial Artificial Chromosome (BAC) screening and shotgun library construction
BAC library of common chimpanzee (Pan troglodytes) RPCI-43 was obtained from BACPAC Resource Center at the Children's Hospital Oakland Research Institute (Oakland, CA). In order to identify BAC clones containing LHB/CGB genome cluster we used recommended protocols and performed hybridization screening with a PCR-product containing LHB-specific sequence amplified from chimpanzee genomic DNA. Probe DNA was labeled with [32P]dCTP by random primer extension using DecaLabel™ DNA Labeling Kit (MBI Fermentas, Vilnius, Lithuania). BAC DNA was isolated using NucleoBond® BAC 100 plasmid purification kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany). Two overlapping BAC clones (68P2 and 109B10) containing LHB/CGB genome cluster were sheared by nebulization to approximately 5 kb long fragments and used for shotgun library construction with TOPO® Shotgun Subcloning Kit (Invitrogen, Carlsbad, CA) according to manufacturers' instructions.
DNA sequencing and data analysis
Plasmid DNA was purified with NucleoSpin®-Plasmid kit (Macherey-Nagel GmbH & Co. KG) and sequenced on ABI 3730 × l sequencer using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Plasmid ends were sequenced using M13F and M13R primers, additional primers for primer walking were designed with the web-based version of the Primer3 software [84]. Sequencing primers are available in Additional file 6. LHB/CGB genome cluster from two chimpanzee BAC clones were sequenced with an average redundancy of 7×, which was sufficient for assembly. Sequences were assembled using ContigExpress program from Vector NTI Suite 9 (Invitrogen) and the chimpanzee sequence was compared to human LHB/CGB genome cluster (GenBank: NG_000019). The full Chimpanzee LHB/CGB cluster sequence has been deposited to GenBank (accession number EU000308). Sequence alignments were performed and homologies determined by the web-based ClustalW [85] and Stretcher implemented in the EMBOSS package [86]. The aligned sequences of the major transcripts of the chimp and human LHB/CGB genes are given in Additional file 7. Substitution and indel divergences were calculated as the percentage of the number of substitutions and the number of nucleotides in indels divided with the total number of aligned nucleotides in the specific genomic region. Phylogenetic trees were constructed by MEGA3.1 [87] using Kimura's two parameter model to infer the neighbor-joining and the branch-and-bound algorithms to find maximum parsimony trees with 1000 replications for bootstrapping.
For coding regions maximum likelihood method [39] was used to estimate non-synonymous/synonymous rate ratio ω (= dn/ds) by CODEML implemented in PAML package version 4 [88, 89]. Codon frequencies were estimated from the dataset using the F3 × 4 option, other settings were as default. The simplest model M0 or one-ratio model was used to estimate the ω (an average over all the sites). As for CGB1 alternative reading frames have been predicted and no functional protein has been characterized so far. We defined CGB1 mRNA sequence and subsequently the predicted reading frame as supported by the published experimental data [18, 90]. In parallel, the number of non-synonymous substitutions per non-synonymous site (dn) and synonymous substitutions per synonymous site (ds) were estimated using an alternative method – the Li93 method [40, 41]. The significance of the difference between dn and ds was examined by a two-tailed Z-test [42] using MEGA3.1. To address which segments of the genes are evolving more rapidly, we performed CRANN analysis [47, 48] using sliding and overlapping windows and for the results in visual documentation of rate heterogeneities of dn and ds, Window size was set on 20 and shift size on 10 codons using the Li93 method.
Repetitive elements were detected by the REPEATMASKER program [91].
Abbreviations
- ADH :
-
alcohol dehydrogenase
- CD33rSiglec :
-
CD33-related Siglecs
- CGB :
-
chorionic gonadotropin beta subunit gene
- FAM90A:
-
family with sequence similarity 90
- GH/CSH:
-
growth hormone/somatomammotropin
- HCG:
-
human chorionic gonadotropin
- KGF:
-
keratinocyte growth factor
- LH:
-
luteinizing hormone: LHB: luteinizing hormone beta subnit gene
- MHC:
-
major histocompatibility complex
- NAIP:
-
neuronal apoptosis inhibitory protein
- NTF5:
-
neurotrophin 5 gene
- PRAME gene family:
-
preferentially expressed antigen of melanoma
- psNTF6:
-
neurotrophin 6 pseudogene(s)
- ROCK1:
-
Rho-dependent protein kinase
- RUVBL2:
-
RuvB-like 2, homologue of the bacterial RuvB gene
- USP10:
-
ubiquitin-specific protease.
References
Ohno S: Evolution by gene duplication. 1970, New York , Springer
Britten RJ: Divergence between samples of chimpanzee and human DNA sequences is 5%, counting indels. Proc Natl Acad Sci U S A. 2002, 99 (21): 13633-13635. 10.1073/pnas.172510699.
Bailey JA, Eichler EE: Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006, 7 (7): 552-564. 10.1038/nrg1895.
Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, Karimpour-Fard A, Glueck D, McGavran L, Berry R, Pollack J, Sikela JM: Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004, 2 (7): E207-10.1371/journal.pbio.0020207.
Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, Rocchi M, Eichler EE: A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005, 437 (7055): 88-93. 10.1038/nature04000.
Britten RJ, Rowen L, Williams J, Cameron RA: Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci U S A. 2003, 100 (8): 4661-4665. 10.1073/pnas.0330964100.
Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y, Fukuzumi Y, Yamazaki M, Tashiro H, Iwamoto C, Umehara Y, Imanishi T, Meyer A, Ikeo K, Gojobori T, Bahram S, Inoko H: Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci U S A. 2003, 100 (13): 7708-7713. 10.1073/pnas.1230533100.
Ebersberger I, Metzler D, Schwarz C, Paabo S: Genomewide comparison of DNA sequences between humans and chimpanzees. Am J Hum Genet. 2002, 70 (6): 1490-1497. 10.1086/340787.
Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK: Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005, 437 (7055): 69-87. 10.1038/nature04072.
Watanabe H, Fujiyama A, Hattori M, Taylor TD, Toyoda A, Kuroki Y, Noguchi H, BenKahla A, Lehrach H, Sudbrak R, Kube M, Taenzer S, Galgoczy P, Platzer M, Scharfe M, Nordsiek G, Blocker H, Hellmann I, Khaitovich P, Paabo S, Reinhardt R, Zheng HJ, Zhang XL, Zhu GF, Wang BF, Fu G, Ren SX, Zhao GP, Chen Z, Lee YS, Cheong JE, Choi SH, Wu KM, Liu TT, Hsiao KJ, Tsai SF, Kim CG, S OO, Kitano T, Kohara Y, Saitou N, Park HS, Wang SY, Yaspo ML, Sakaki Y: DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature. 2004, 429 (6990): 382-388. 10.1038/nature02564.
Wetterbom A, Sevov M, Cavelier L, Bergstrom TF: Comparative genomic analysis of human and chimpanzee indicates a key role for indels in primate evolution. J Mol Evol. 2006, 63 (5): 682-690. 10.1007/s00239-006-0045-7.
Chen FC, Vallender EJ, Wang H, Tzeng CS, Li WH: Genomic divergence between human and chimpanzee estimated from large-scale alignments of genomic sequences. J Hered. 2001, 92 (6): 481-489. 10.1093/jhered/92.6.481.
Stankiewicz P, Lupski JR: Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002, 18 (2): 74-82. 10.1016/S0168-9525(02)02592-1.
Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, Oseroff VV, Albertson DG, Pinkel D, Eichler EE: Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005, 77 (1): 78-88. 10.1086/431652.
Li MD, Ford JJ: A comprehensive evolutionary analysis based on nucleotide and amino acid sequences of the alpha- and beta-subunits of glycoprotein hormone gene family. J Endocrinol. 1998, 156 (3): 529-542. 10.1677/joe.0.1560529.
Maston GA, Ruvolo M: Chorionic gonadotropin has a recent origin within primates and an evolutionary history of selection. Mol Biol Evol. 2002, 19 (3): 320-335.
Freitas EM, Gaudieri S, Zhang WJ, Kulski JK, van Bockxmeer FM, Christiansen FT, Dawkins RL: Duplication and diversification of the apolipoprotein CI (APOCI) genomic segment in association with retroelements. J Mol Evol. 2000, 50 (4): 391-396.
Bo M, Boime I: Identification of the transcriptionally active genes of the chorionic gonadotropin beta gene cluster in vivo. J Biol Chem. 1992, 267 (5): 3179-3184.
Hollenberg AN, Pestell RG, Albanese C, Boers ME, Jameson JL: Multiple promoter elements in the human chorionic gonadotropin beta subunit genes distinguish their expression from the luteinizing hormone beta gene. Mol Cell Endocrinol. 1994, 106 (1-2): 111-119. 10.1016/0303-7207(94)90192-9.
Li Y, Ye C, Shi P, Zou XJ, Xiao R, Gong YY, Zhang YP: Independent origin of the growth hormone gene family in New World monkeys and Old World monkeys/hominoids. J Mol Endocrinol. 2005, 35 (2): 399-409. 10.1677/jme.1.01778.
Revol De Mendoza A, Esquivel Escobedo D, Martinez Davila I, Saldana H: Expansion and divergence of the GH locus between spider monkey and chimpanzee. Gene. 2004, 336 (2): 185-193. 10.1016/j.gene.2004.03.034.
Hunt DM, Dulai KS, Cowing JA, Julliot C, Mollon JD, Bowmaker JK, Li WH, Hewett-Emmett D: Molecular evolution of trichromacy in primates. Vision Res. 1998, 38 (21): 3299-3306. 10.1016/S0042-6989(97)00443-4.
Kainz PM, Neitz J, Neitz M: Recent evolution of uniform trichromacy in a New World monkey. Vision Res. 1998, 38 (21): 3315-3320. 10.1016/S0042-6989(98)00078-9.
Dulai KS, von Dornum M, Mollon JD, Hunt DM: The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Res. 1999, 9 (7): 629-638.
Wallis OC, Wallis M: Characterisation of the GH gene cluster in a new-world monkey, the marmoset (Callithrix jacchus). J Mol Endocrinol. 2002, 29 (1): 89-97. 10.1677/jme.0.0290089.
Chen FC, Li WH: Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001, 68 (2): 444-456. 10.1086/318206.
Batzer MA, Kilroy GE, Richard PE, Shaikh TH, Desselle TD, Hoppens CL, Deininger PL: Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 1990, 18 (23): 6793-6798. 10.1093/nar/18.23.6793.
Kudla G, Helwak A, Lipinski L: Gene conversion and GC-content evolution in mammalian Hsp70. Mol Biol Evol. 2004, 21 (7): 1438-1444. 10.1093/molbev/msh146.
Hallast P, Nagirnaja L, Margus T, Laan M: Segmental duplications and gene conversion: Human luteinizing hormone/chorionic gonadotropin beta gene cluster. Genome Res. 2005, 15 (11): 1535-1546. 10.1101/gr.4270505.
Galtier N, Duret L: Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007, 23 (6): 273-277. 10.1016/j.tig.2007.03.011.
Marais G: Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003, 19 (6): 330-338. 10.1016/S0168-9525(03)00116-1.
Nachman MW, Crowell SL: Estimate of the mutation rate per nucleotide in humans. Genetics. 2000, 156 (1): 297-304.
Shi J, Xi H, Wang Y, Zhang C, Jiang Z, Zhang K, Shen Y, Jin L, Zhang K, Yuan W, Wang Y, Lin J, Hua Q, Wang F, Xu S, Ren S, Xu S, Zhao G, Chen Z, Jin L, Huang W: Divergence of the genes on human chromosome 21 between human and other hominoids and variation of substitution rates among transcription units. Proc Natl Acad Sci U S A. 2003, 100 (14): 8331-8336. 10.1073/pnas.1332748100.
Wildman DE, Uddin M, Liu G, Grossman LI, Goodman M: Implications of natural selection in shaping 99.4% nonsynonymous DNA identity between humans and chimpanzees: enlarging genus Homo. Proc Natl Acad Sci U S A. 2003, 100 (12): 7181-7188. 10.1073/pnas.1232172100.
Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, Wilson RK, Page DC: Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005, 437 (7055): 100-103. 10.1038/nature04101.
Elango N, Thomas JW, Yi SV: Variable molecular clocks in hominoids. Proc Natl Acad Sci U S A. 2006, 103 (5): 1370-1375. 10.1073/pnas.0510716103.
Walsh JB: Interaction of selection and biased gene conversion in a multigene family. Proc Natl Acad Sci U S A. 1985, 82 (1): 153-157. 10.1073/pnas.82.1.153.
Ohta T: Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proc Natl Acad Sci U S A. 1991, 88 (15): 6716-6720. 10.1073/pnas.88.15.6716.
Goldman N, Yang Z: A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994, 11 (5): 725-736.
Li WH: Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993, 36 (1): 96-99. 10.1007/BF02407308.
Pamilo P, Bianchi NO: Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol Biol Evol. 1993, 10 (2): 271-281.
Nei M, Kumar S: Molecular Evolution and Phylogenetics. 2000, New York , OXFORD University press
Rull K, Laan M: Expression of beta-subunit of HCG genes during normal and failed pregnancy. Hum Reprod. 2005, 20 (12): 3360-3368. 10.1093/humrep/dei261.
Hallast P, Rull K, Laan M: The evolution and genomic landscape of CGB1 and CGB2 genes. Mol Cell Endocrinol. 2007, 260-262: 2-11. 10.1016/j.mce.2005.11.049.
Rull K, Hallast P, Uuskula L, Jackson J, Punab M, Salumets A, Campbell RK, Laan M: Fine-scale quantification of HCG beta gene transcription in human trophoblastic and non-malignant non-trophoblastic tissues. Mol Hum Reprod. 2008, 14 (1): 23-31. 10.1093/molehr/gam082.
Berger P, Gruschwitz M, Spoettl G, Dirnhofer S, Madersbacher S, Gerth R, Merz WE, Plas E, Sampson N: Human chorionic gonadotropin (hCG) in the male reproductive tract. Mol Cell Endocrinol. 2007, 260-262: 190-196. 10.1016/j.mce.2006.01.021.
Creevey CJ, McInerney JO: An algorithm for detecting directional and non-directional positive selection, neutrality and negative selection in protein coding DNA sequences. Gene. 2002, 300 (1-2): 43-51. 10.1016/S0378-1119(02)01039-9.
Creevey CJ, McInerney JO: CRANN: detecting adaptive evolution in protein-coding DNA sequences. Bioinformatics. 2003, 19 (13): 1726-10.1093/bioinformatics/btg225.
Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, Pollack JR, Sikela JM: Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007, 17 (9): 1266-1277. 10.1101/gr.6557307.
Piontkivska H, Nei M: Birth-and-death evolution in primate MHC class I genes: divergence time estimates. Mol Biol Evol. 2003, 20 (4): 601-609. 10.1093/molbev/msg064.
Semple CA, Rolfe M, Dorin JR: Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol. 2003, 4 (5): R31-10.1186/gb-2003-4-5-r31.
Angata T, Margulies EH, Green ED, Varki A: Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci U S A. 2004, 101 (36): 13251-13256. 10.1073/pnas.0404833101.
Birtle Z, Goodstadt L, Ponting C: Duplication and positive selection among hominin-specific PRAME genes. BMC Genomics. 2005, 6: 120-10.1186/1471-2164-6-120.
Fitch DH, Bailey WJ, Tagle DA, Goodman M, Sieu L, Slightom JL: Duplication of the gamma-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci U S A. 1991, 88 (16): 7396-7400. 10.1073/pnas.88.16.7396.
Salvignol I, Blancher A, Calvas P, Socha WW, Colin Y, Cartron JP, Ruffie J: Relationship between chimpanzee Rh-like genes and the R-C-E-F blood group system. J Med Primatol. 1993, 22 (1): 19-28.
Oota H, Dunn CW, Speed WC, Pakstis AJ, Palmatier MA, Kidd JR, Kidd KK: Conservative evolution in duplicated genes of the primate Class I ADH cluster. Gene. 2007, 392 (1-2): 64-76. 10.1016/j.gene.2006.11.008.
Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD: Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007, 7: 187-10.1186/1471-2148-7-187.
Bosch N, Caceres M, Cardone MF, Carreras A, Ballana E, Rocchi M, Armengol L, Estivill X: Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum Mol Genet. 2007, 16 (21): 2572-2582. 10.1093/hmg/ddm209.
Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM: Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006, 313 (5791): 1304-1307. 10.1126/science.1127980.
Bailey JA, Liu G, Eichler EE: An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003, 73 (4): 823-834. 10.1086/378594.
Chen EY, Liao YC, Smith DH, Barrera-Saldana HA, Gelinas RE, Seeburg PH: The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics. 1989, 4 (4): 479-497. 10.1016/0888-7543(89)90271-1.
Ye C, Li Y, Shi P, Zhang YP: Molecular evolution of growth hormone gene family in old world monkeys and hominoids. Gene. 2005, 350 (2): 183-192. 10.1016/j.gene.2005.03.003.
Golos TG, Durning M, Fisher JM, Fowler PD: Cloning of four growth hormone/chorionic somatomammotropin-related complementary deoxyribonucleic acids differentially expressed during pregnancy in the rhesus monkey placenta. Endocrinology. 1993, 133 (4): 1744-1752. 10.1210/en.133.4.1744.
Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan-Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour-Fard A, Kim YH, Pollack JR, Vinar T, Addo-Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer-Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O'Brien W E, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS: Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007, 316 (5822): 222-234. 10.1126/science.1139247.
Boyson JE, Shufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Hughes AL, Watkins DI: The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996, 156 (12): 4656-4665.
Chen ZW, McAdam SN, Hughes AL, Dogon AL, Letvin NL, Watkins DI: Molecular cloning of orangutan and gibbon MHC class I cDNA. The HLA-A and -B loci diverged over 30 million years ago. J Immunol. 1992, 148 (8): 2547-2554.
Cooper GM, Nickerson DA, Eichler EE: Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 2007, 39 (7 Suppl): S22-9. 10.1038/ng2054.
Ciccodicola A, D'Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franze A, Cuccurese M, Cocchia M, Curci A, Terracciano A, Torino A, Cocchia S, Mercadante G, Pannone E, Archidiacono N, Rocchi M, Schlessinger D, D'Urso M: Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000, 9 (3): 395-401. 10.1093/hmg/9.3.395.
Zimonjic DB, Kelley MJ, Rubin JS, Aaronson SA, Popescu NC: Fluorescence in situ hybridization analysis of keratinocyte growth factor gene amplification and dispersion in evolution of great apes and humans. Proc Natl Acad Sci U S A. 1997, 94 (21): 11461-11465. 10.1073/pnas.94.21.11461.
Fairbanks DJ, Maughan PJ: Evolution of the NANOG pseudogene family in the human and chimpanzee genomes. BMC Evol Biol. 2006, 6: 12-10.1186/1471-2148-6-12.
Hughes AL, Nei M: Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988, 335 (6186): 167-170. 10.1038/335167a0.
Aguileta G, Bielawski JP, Yang Z: Gene conversion and functional divergence in the beta-globin gene family. J Mol Evol. 2004, 59 (2): 177-189. 10.1007/s00239-004-2612-0.
Aguileta G, Bielawski JP, Yang Z: Evolutionary rate variation among vertebrate beta globin genes: implications for dating gene family duplication events. Gene. 2006, 380 (1): 21-29. 10.1016/j.gene.2006.04.019.
de Groot NG, Otting N, Doxiadis GG, Balla-Jhagjhoorsingh SS, Heeney JL, van Rood JJ, Gagneux P, Bontrop RE: Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci U S A. 2002, 99 (18): 11748-11753. 10.1073/pnas.182420799.
de Groot NG, Garcia CA, Verschoor EJ, Doxiadis GG, Marsh SG, Otting N, Bontrop RE: Reduced MIC gene repertoire variation in West African chimpanzees as compared to humans. Mol Biol Evol. 2005, 22 (6): 1375-1385. 10.1093/molbev/msi127.
Simula AP, Amato F, Faast R, Lopata A, Berka J, Norman RJ: Luteinizing hormone/chorionic gonadotropin bioactivity in the common marmoset (Callithrix jacchus) is due to a chorionic gonadotropin molecule with a structure intermediate between human chorionic gonadotropin and human luteinizing hormone. Biol Reprod. 1995, 53 (2): 380-389. 10.1095/biolreprod53.2.380.
Muller T, Simoni M, Pekel E, Luetjens CM, Chandolia R, Amato F, Norman RJ, Gromoll J: Chorionic gonadotrophin beta subunit mRNA but not luteinising hormone beta subunit mRNA is expressed in the pituitary of the common marmoset (Callithrix jacchus). J Mol Endocrinol. 2004, 32 (1): 115-128. 10.1677/jme.0.0320115.
Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP: Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007, 8 (10): 762-775. 10.1038/nrg2193.
Cheung B, Holmes RS, Easteal S, Beacham IR: Evolution of class I alcohol dehydrogenase genes in catarrhine primates: gene conversion, substitution rates, and gene regulation. Mol Biol Evol. 1999, 16 (1): 23-36.
Wallis OC, Wallis M: Evolution of growth hormone in primates: the GH gene clusters of the New World monkeys marmoset (Callithrix jacchus) and white-fronted capuchin (Cebus albifrons). J Mol Evol. 2006, 63 (5): 591-601. 10.1007/s00239-006-0039-5.
Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, Anzai T, Kulski JK, Kikkawa E, Naruse T, Kimura N, Yanagiya K, Watanabe A, Hosomichi K, Kohara S, Iwamoto C, Umehara Y, Meyer A, Wanner V, Sano K, Macquin C, Ikeo K, Tokunaga K, Gojobori T, Inoko H, Bahram S: Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006, 173 (3): 1555-1570. 10.1534/genetics.106.057034.
Esteban C, Audi L, Carrascosa A, Fernandez-Cancio M, Perez-Arroyo A, Ulied A, Andaluz P, Arjona R, Albisu M, Clemente M, Gussinye M, Yeste D: Human growth hormone (GH1) gene polymorphism map in a normal-statured adult population. Clin Endocrinol (Oxf). 2007, 66 (2): 258-268. 10.1111/j.1365-2265.2006.02718.x.
Sedman L, Padhukasahasram B, Kelgo P, Laan M: Complex signatures of locus-specific selective pressures and gene conversion on Human Growth Hormone/Chorionic Somatomammotropin genes. Hum Mutat. 2008
Primer3 software. [http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi]
ClustalW. [http://www.ebi.ac.uk/clustalw/]
EMBOSS package. [http://emboss.sourceforge.net/]
Kumar S, Tamura K, Nei M: MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004, 5 (2): 150-163. 10.1093/bib/5.2.150.
Yang Z: PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997, 13 (5): 555-556.
Yang Z: PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007, 24 (8): 1586-1591. 10.1093/molbev/msm088.
Dirnhofer S, Hermann M, Hittmair A, Hoermann R, Kapelari K, Berger P: Expression of the human chorionic gonadotropin-beta gene cluster in human pituitaries and alternate use of exon 1. J Clin Endocrinol Metab. 1996, 81 (12): 4212-4217. 10.1210/jc.81.12.4212.
REPEATMASKER program. [http://www.repeatmasker.org/]
Acknowledgements
We thank Tõnu Margus, Siim Sõber, Tarmo Annilo, Pekka Ellonen, Mari Kaunisto, Maija Wessman and Verneri Anttila for discussions and advice, and Kärt Tomberg for editing the English language. M.L. is a Wellcome Trust International Senior Research Fellow (grants no. 070191/Z/03/Z) in Biomedical Science in Central Europe and a HHMI International Scholar (grant #55005617). Additionally, the study has been supported by the Estonian Ministry of Education and Science core grant no. 0182721s06 and the Estonian Science Foundation grant no. 5796 (M.L., P.H.), as well as personal scholarships from the Centre for International Mobility (CIMO), Kristjan Jaak Stipend Program and World Federation of Scientists' (P.H.), the Center of Excellence grant of Complex Disease Genetics of the Academy of Finland and the Sigrid Juselius Foundation (J.S., A.P).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
PH, JS, AP and ML designed the research; PH performed the research; JS, AP contributed to the analytic tools; PH and ML analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
12862_2007_764_MOESM1_ESM.pdf
Additional file 1: Table for sequence parameters. Sequence parameters for the intergenic regions and for the whole LHB/CGB cluster in human and chimpanzee. (PDF 50 KB)
12862_2007_764_MOESM2_ESM.pdf
Additional file 2: Distribution of species-specific indels. For simplification we defined all identified gaps in sequence alignments of human and chimpanzee LHB/CGB region orthologous segments (Figure 1B) as deletions in one of the species. The figure shows the number of species-specific gaps (Y-axes) relative to their length in base pairs (X-axis) and the contribution of each deletion class (1–5; 6–10; 11–15; 16–20; 20–25; >100 bp) to the total length of species-specific gapped sequence. (PDF 49 KB)
12862_2007_764_MOESM3_ESM.pdf
Additional file 3: Table for nucleotide sequence divergence. Nucleotide sequence divergence in orthologous human and chimpanzee LHB/CGB genes. Divergence was estimated by using the human (GenBank: NG_000019) and chimpanzee (this study, Genbank: EU000308) reference sequences alone or by incorporating the re-sequencing data for one or both species (for human n = 95 [29]; for chimpanzee, n = 11, unpublished data of the authors) into the calculations. (PDF 73 KB)
12862_2007_764_MOESM4_ESM.pdf
Additional file 4: Table of estimated dn and ds values by Li93 method. Application of Li93 method [40, 41] for estimating non-synonymous (dn) and synonymous (ds) substitutions, and amino acid divergence in human and chimpanzee orthologous genes and for testing significance in the deviation of dn/ds ratio from expectation under neutrality. (PDF 44 KB)
12862_2007_764_MOESM5_ESM.pdf
Additional file 5: Results of the CRANN analyses of human and chimpanzee orthologous genes (A) LHB, (B) CGB5, (C) CGB8, (D) CGB7, (E) CGB1. Results of moving window analysis carried out with CRANN [47, 48]. X-axis shows the successive windows of 20 codon sites (window size: 20 codons, shift size: 10 codons). As the number of substitutions calculated in each moving window for human and chimpanzee orthologous genes was low, the dn and ds values were mostly zero and thus the dn/ds ratio was not shown. (PDF 184 KB)
12862_2007_764_MOESM6_ESM.pdf
Additional file 6: Primer sequences. Primers for sequencing of subcloned BACs 68P2 and 109B10 originating from common chimpanzee (Pan troglodytes) BAC library RPCI-43 (BACPAC Resource Center at the Children's Hospital Oakland Research Institute; Oakland, CA). (PDF 61 KB)
12862_2007_764_MOESM7_ESM.pdf
Additional file 7: Alignments of the chimp and human LHB/CGB transcripts. The aligned sequences of the major transcripts of the chimp and human LHB/CGB genes. The ATG of each gene has been indicated in red and underlined font. Translation STOP codons have been boxed. (PDF 26 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hallast, P., Saarela, J., Palotie, A. et al. High divergence in primate-specific duplicated regions: Human and chimpanzee Chorionic Gonadotropin Betagenes. BMC Evol Biol 8, 195 (2008). https://doi.org/10.1186/1471-2148-8-195
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-8-195