Abstract
Chronic obstructive pulmonary disease (COPD) is a treatable and preventable disease state, characterised by progressive airflow limitation that is not fully reversible. Although COPD is primarily a disease of the lungs there is now an appreciation that many of the manifestations of disease are outside the lung, leading to the notion that COPD is a systemic disease. Currently, diagnosis of COPD relies on largely descriptive measures to enable classification, such as symptoms and lung function. Here the limitations of existing diagnostic strategies of COPD are discussed and systems biology approaches to diagnosis that build upon current molecular knowledge of the disease are described. These approaches rely on new 'label-free' sensing technologies, such as high-throughput surface plasmon resonance (SPR), that we also describe.
Similar content being viewed by others
Chronic Obstructive Pulmonary Disease
Chronic obstructive pulmonary disease (COPD) is a treatable and preventable condition characterised by progressive airflow limitation that is not fully reversible [1]. COPD is associated with an abnormal inflammatory response of the lungs to noxious particles or gases. This is primarily caused by tobacco smoking [2, 3] but there is gathering evidence that additional factors predispose patients to COPD, such as genetic susceptibility, air pollution and other airborne irritants [4, 5]. There may well be a genetic predisposition and also some food preservatives have also been implicated indicating that the underlying causality of the disease may not just reside in lung insult from the atmosphere [6]. COPD is projected to have a major effect on human health and worldwide by 2020 it is predicted to be the third most frequent cause of death [7].
COPD consists of three main respiratory pathologies; emphysema, respiratory bronchiolitis and chronic bronchitis. These separate and distinct pathologies illustrate the heterogeneity of COPD [8] and the importance of well defined COPD phenotypes [9]. Although COPD is primarily a disease of the lungs there is now an appreciation that many of the manifestations of disease are outside the lung, such as cachexia, skeletal muscle dysfunction, cardiovascular disease, depression and osteoporosis [10], leading to the concept that COPD is a systemic disease [11–15].
Current Methods for Confirming a COPD Diagnosis
The diagnosis of COPD is based on the presence of typical symptoms of cough and shortness of breath, together with the presence of risk factors, and is confirmed by spirometry. A variety of methods (as outlined in Figure 1) are then used to classify the severity of disease, including questionnaires, GOLD and BODE Index.
The Global Initiative for Chronic Obstructive Lung Disease (GOLD) classifies COPD into four stages; mild, moderate, severe and very severe according to spirometric measurements [16]. Spirometry, however, is believed to correlate poorly with symptoms [17], quality of life [18], exacerbation frequency [19] and exercise intolerance [20].
A more recent and comprehensive method for assessing disease severity and prognosis of COPD is the BODE Index. This is a multidimensional grading system, which not only measures airflow obstruction (FEV1), but also incorporates body mass index (BMI), dyspnoea score and exercise capacity [21]. A comparison between the BODE and GOLD classifications shows that the BODE is a better predictor of hospitalisation [22] and death [21] than by GOLD.
There are conflicting views on the prevalence of COPD ranging from 3–12% [23] to 50% [24]. A major contributing factor to this may be that only one-third of physicians know the correct spirometric criteria according to GOLD [25] and only one-third of trained GPs and nurses trust their own spirometric interpretive skills [26]. Additionally, the technical limitations of the instruments used to undertake these spirometric measurements such as instrument variation and signal-to-noise ratio need to be considered [27, 28]. Although spirometry is generally used to measure airflow obstruction, it has a number of limitations with regard to the detection and assessment of disease. Spirometry measures established airflow obstruction, which is likely to result from a long and continuous inflammatory process. Early use of therapeutic interventions, however, may be most helpful in attenuating the development of airway obstruction, which is not identifiable by spirometric tests. A single FEV1 measurement will give information on how much airway obstruction has already occurred, but will not give any information as to the current level of disease activity. At present, such information can only be obtained by serial measurements and assessment of the reduction in FEV1 over time. Finally, spirometry measures the end result of what may be a number of disease processes. It is known that patients vary considerably in their response to treatments, for example to inhaled corticosteroids [29], and it is possible that there are a number of pathways by which smoking and other exposures lead to the final state of COPD. An alternative diagnostic approach may help identify disease subtypes and allow for a more accurate distinction between COPD and chronic irreversible asthma [30].
Biomarker Identification
In an effort to identify biomarkers of COPD, several groups have looked at genetic susceptibility (single nucleotide polymorphisms; SNPs), gene expression or protein expression. The observations from these studies have provided useful information and insights into the pathogenesis of COPD.
Genetic susceptibility
As previously mentioned, COPD is associated with an abnormal inflammatory response of the lungs to noxious particles or gases. Due to the diverse response to these environmental insults, it is likely that genetic factors are important within the aetiology of COPD [31], but only severe alpha 1-antitrypsin deficiency is a proven genetic risk factor for COPD [32].
To date, studies have taken one of two approaches; they have either focused on candidate genes such as CCL5 [33] or taken a more holistic approach and completed genome-wide linkage analysis studies to identify regions of the genome that confer susceptibility [34]. The major considerations with any genetic study, however, are the large size required and the need for replication in a different, large data set. Using the focused approach Chappell et al have identified six haplotypes of the SERPINA1 gene that increases the risk of disease [35]. A recent genome-wide linkage analysis performed by Hersh et al identified a region on chromosome 1p that showed strong evidence of linkage to lung function traits [36]. Association analysis then identified TGFBR3 (betaglycan) as a potential susceptibility gene for COPD, which is supported by both murine and human microarray data.
Gene Expression
Several researchers have examined gene expression profiles in an attempt to identify biomarkers, distinguish disease sub-types and generate candidates for further genetic and biological studies [37–45].
Spira et al reported genome-wide expression profiling of subjects with severe emphysema undergoing lung volume reduction surgery, which identified gene expression markers for severe emphysema as well as positive response to surgery [44]. Golpon et al used a similar approach and identified gene expression biomarkers distinguishing patients with α1-antitrypsin deficiency [41]. Pierrou and colleagues have identified a gene set of 200 transcripts dysregulated in COPD compared to healthy smokers [37]. As with most disease-focused microarray studies, however, there has been a lack of consistency in the identification of COPD gene expression biomarkers. For example, Egr-1 was identified in a microarray study as a gene over-expressed in emphysema subjects by Zhang et al [46]. Subsequently, Ning et al, using a combined microarray/SAGE approach, validated Egr-1 induction associated with COPD severity [40]. Ning et al went on to show that Egr-1 appears to contribute to disease pathogenesis, as it can regulate matrix-remodelling potential through fibroblast protease production. Bhattacharya et al, however, found no evidence of differential expression for Egr-1 in their population, although this study is one of the most promising to date, as the authors have presented the first gene expression biomarker for COPD validated in an independent data set [45]. This study, however, still has limitations, mainly due to the size of the sample population.
Overall, there is minimal overlap between differentially expressed genes among the different datasets. This problem highlights the complexity of expression profiling analysis in a human disease, such as COPD, with tissue heterogeneity and variable clinical phenotype. The non-overlapping gene datasets from these studies are due to several factors, including differences in sample acquisition, disease severity, sample size, tissue and cell components, and expression platforms [39].
Protein Expression
Numerous groups have looked at protein expression, but most studies, due to technology limitations, have only analysed a limited set of proteins [47–52]. Shaker and colleagues examined six plasma proteins of known potential interest in COPD by enzyme-linked immunosorbant assay (ELISA) [48]. From this extremely selective reductionist approach they were able to show that some proteins were up-regulated and some were down-regulated, which emphasises the need for a more holistic approach to deliver a molecular fingerprint of disease. A larger scale analysis of proteins in COPD has been undertaken using two different techniques. Plymoth et al, by using a combination of replicate 2-dimensional gel separations, image annotation, and mass spectrometry identification, were able to investigate 406 proteins in bronchoalveolar lavage (BAL) that had the potential to identify smokers at risk of developing COPD [49]. These proteins showed expression patterns that were both up- and down-regulated. Pinto-Plata et al went a stage further and used serum on a 'Protein Microarray Platform' (PMP), which provided data on 143 serum proteins of potential interest [50]. This highlighted 24 proteins, which were up-regulated in disease, but it was acknowledged by the authors that the study was a proof of principle rather than a comprehensive analysis of all possible biomolecules related to COPD.
Systems Biology: A New Approach to Disease Diagnosis and Management
Despite intensive research, definitive single disease-defining biomarkers for COPD remain elusive. Molecules shown to have a significant correlation with disease status often fail to accurately discriminate COPD from closely related diseases that display similar symptoms. As such, many of the potential biomarkers that have been suggested for COPD, including proteins [50, 51, 53], cytokines [48, 50, 54–65], antibodies [66], enzymes [50, 67–69] and inhibitors [48], have also been implicated as potential targets in other lung diseases or general systemic inflammation [70–116] (Table 1). The difficulties encountered whilst searching for COPD biomarkers may be due in part to the complex nature of the disease, which comprises a broad spectrum of histopathological findings and respiratory symptoms [45]. Consequently, the probability of finding a single marker that is representative of all these processes is rather unlikely. Identification of single biomarkers is also hindered by the high level of variability in normal protein concentrations amongst individuals. This makes it difficult to establish the concentration of a single mediator that indicates disease onset [117, 118]. Thus, it is essential to put isolated readings into context, i.e., does an elevated protein concentration indicate the presence of disease, or is it just a high but otherwise normal reading?
The problems encountered with biomarker identification are not unique to COPD. Whilst the focus of biomarker studies over the last decade or so has primarily been placed on the use of individual molecular biomarkers as indicators of disease, this approach has only proved successful for a limited number of diseases including prostate and breast cancer where measurements of prostate specific antigen (PSA) and human epidermal growth factor receptor 2 (HER2) respectively are routinely used in diagnostic procedures [119, 120]. New approaches to disease diagnosis in general, therefore, are required.
Systems biology is a broad new paradigm that has recently entered the terminology of the life and biomedical sciences arena. It is an integrative approach focused on deciphering the relationship and the interactions between the gene, protein and cell elements of a biological system and how they impact on the function and behaviour of that system [121] (Figure 2). Traditional '-omics' fields, including genomics, proteomics, metabolomics and transcriptomics examine only one strand of the information available about an organism. Systems biology combines data from all these fields with bioinformatic, computational biology and engineering principles to examine organisms as systems of interconnecting networks. These networks will be modelled according to initial data obtained by traditional '-omics' and then revised through a combination of iterative refinement and bootstrapping (repeated random samples taken from a dataset) as described by Aderem [122] and Lucas [123]. By studying complex biological systems in this way, it is possible to identify emergent properties that are not demonstrated by individual '-omics' fields and cannot be predicted even with full understanding of the parts alone. A comprehensive understanding of these emergent properties requires systems-level perspectives not obtainable using simple reductionist approaches [122].
Studies have started to apply systems biology approaches and principles to decipher the pathways underlying complex diseases including Alzheimer's disease [124], polyarticular juvenile idiopathic arthritis [125], psychiatric disorders [126] and Sjögren's syndrome [127]. Application of the integrative approach provided by systems biology seems to offer a better route to understanding disease [128, 129]. Currently, our understanding of systems biology is reaching a point whereby patterns of molecular behaviour are far clearer indicators of pathophysiological conditions than individual molecular markers [129]. Each disease possesses a unique molecular fingerprint that could be used diagnostically to differentiate it from diseases with closely related phenotypes. This novel concept, whilst still in its infancy, is being applied to cancer diagnosis [130] and is ideal for diagnosis of other complex diseases such as COPD.
Identification of a COPD-specific molecular fingerprint is a sizeable problem due to the heterogeneity of the disease and represents a huge undertaking. Different disease subtypes would each display slight, but measureable, variations of an overall COPD fingerprint. This fingerprint would also need to be sensitive enough to discriminate between COPD and other respiratory diseases e.g. chronic asthma, many of which display similar symptoms.
Initially, the COPD-specific molecular fingerprint would comprise biomolecules already associated with the disease, such as the RNA and protein molecules previously mentioned. Whilst these are the most well characterised disease targets, other molecular species may eventually form an integral part of a disease-specific molecular fingerprint. Targets such as SNPs [131], miRNA [132, 133] and post-translational modifications [134, 135] have all been shown to be important in disease pathology. Thus, a disease-specific molecular fingerprint would be a dynamic model that could be adapted to include such targets as new evidence becomes available of their involvement in COPD.
Current Analytical Technologies
The feasibility of identifying disease-specific biomolecular patterns has been enhanced by the recent advent of proteomic and genomic technologies. Multi-parametric technologies, including bead-based assays (i.e., Luminex and Cytokine Bead Arrays), 2D gel electrophoresis, microarray platforms (both DNA and protein) and mass spectrometry, have provided the opportunities for a more holistic approach not previously possible using conventional technologies such as the enzyme-linked immunosorbent assay (ELISA) [136–140]. The implementation of these high-throughput technologies has vastly increased the prospects of biomarker research as they facilitate simultaneous analysis of multiple (often tens of thousands) potential biomarkers in minimal sample volumes with the potential for identifying novel targets not previously associated with the disease of interest. As such, they will be vital during the extremely complex task of identifying and revising disease-specific molecular fingerprints. Employment of systems biology approaches in routine diagnostic procedures, however, would require the availability of technologies that allow simultaneous detection of different molecular species e.g. both genes and proteins. The major disadvantage with the aforementioned techniques is the ability to detect only a single molecular species at once. Limitations with traditional proteomic and genomic technologies, particularly ELISA- and fluorescence-based systems, would be prohibitive to the production of systems that simultaneously detect multiple types of biomolecule. Such difficulties, including reagent limitations, the need for lengthy and complicated labeling, incubation and detection procedures and the potential for steric hindrance caused by the label at the binding site, could all be circumvented by the use of label-free technologies [141–143] such as surface plasmon resonance (SPR).
Surface Plasmon Resonance (SPR)
What is SPR?
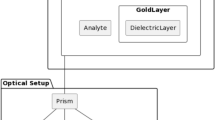
Surface plasmon resonance (SPR) polaritons are surface electromagnetic waves that propagate in a direction parallel to the interface between the metal surface and the external medium e.g., liquid. Since the wave exists on the boundary of the metal and the external liquid medium, these oscillations are very sensitive to any change of this boundary, such as the adsorption of molecules to the metal surface. This phenomenon enables the label-free, real-time detection of the interaction of biological molecules to the metal surface (usually gold) [144]. One frequently used configuration of the technology comprises a glass surface, coated with a thin gold film, which is attached to a prism (Figure 3). Chemical modification of the gold surface allows for the attachment of ligands for many different biomolecules [145–148]. Polarized light from a laser or other light source interacts with the gold surface at an angle greater than the critical angle (θ). Above this angle the light is coupled to electrons in the gold surface resulting in the propagation of surface plasmons along the surface. A surface plasmon only penetrates a short distance into the external medium (e.g., the aqueous environment in a flow cell) making it highly sensitive to changes on the surface of the gold but largely unaffected by processes in the bulk medium. Changes on the surface due to binding events can be readily monitored and have the potential to be used to measure concentrations, ligand-receptor binding affinities and association-dissociation kinetics of potentially thousands of proteins and genes rapidly and simultaneously [143].
Outline of a Surface Plasmon Resonance (SPR) system utilising a Kretschman-Raether configuration. A system with this configuration facilitates label-free detection of biomolecules that bind in real-time. Biomolecules within the sample bind to ligands immobilised on the gold surface causing a change in the levels of the surface plasmon signals. Analysis of this change enables determination of both kinetic and analyte concentrations.
The use of SPR for the detection of biomolecules
The single great virtue of using SPR-based detection modalities is that they are label-free and thus do not require anything more for their identification apart from selective recognition on an appropriate chip surface. Coupling the appropriate surface chemistry for ligand attachment with SPR would allow detection of virtually any species of biomolecule. If the correct capture molecule is selected, SPR is specific enough to distinguish between different glycosylated forms of an antibody [149]. This flexibility, coupled with the potential for increased sensitivity [150], has led to an upsurge in the use of SPR technology. SPR has traditionally been used for identification of protein binding partners and characterisation of binding events [151–156]. It has been applied to the discovery and development of potential therapeutic agents [157–159] and characterisation of interactions between these compounds and their targets [160, 161]. Additionally, it has been used to characterise the molecules, biochemical interactions and processes that may play a role in disease pathology [162–165].
More recently SPR has emerged as a powerful platform for biomarker studies and has been employed in the measurement of many biomolecules implicated in disease (Table 2). SPR detection systems have now been deployed in assays for a wide range of biomolecular species including proteins [166–172], antibodies [173], SNPs [174], sugars [175, 176], narcotics [177, 178], peptides [179, 180], small molecules [181] and microRNAs [182]. These biomarkers have been identified within multiple types of clinical sample including mock samples [183], plasma [173, 184–188], serum [189] and saliva [181, 190]. Several of the studies mentioned in Table 2 have used SPR to detect biomarkers at clinically relevant concentrations highlighting the feasibility of using SPR in a clinical setting. For example, Nagel et al have been able to differentiate Lyme borreliosis infected patients from healthy donors by SPR analysis of Lyme borreliosis specific antibodies in blood serum samples [188]. Cho et al used SPR detection of CSFV antibodies to identify pigs infected with classical swine fever [191]. Vaisocherova et al devised an SPR assay for detection of the candidate pancreatic cancer marker activated cell leukocyte adhesion molecule (ALCAM) that can be used to distinguish between ALCAM levels in cancer and control sera [192]. The measurements made during the latter two studies were demonstrated to have comparative specificity and sensitivity to those undertaken with classical detection techniques [191, 192]. SPR, however, has the additional benefits of being label-free, requiring no amplification step, having low sample requirement and high reusability, and requiring no sample pretreatment [192, 193]. These advantages will in turn result in decreased experimental time, increased cost efficiency and simplification of detection protocols allowing lower user proficiency.
Systems Biology Approaches to COPD Diagnosis – Implementation of a working COPD specific microarray chip
The principles of SPR, when combined with the use of an imaging step (SPR imaging; SPRi), allows a gold surface to be prepared in an array format providing the opportunity to study thousands of interactions rapidly and simultaneously [194]. SPRi could be employed in the development of a COPD specific microarray chip onto which ligands to the biomolecular components of the COPD-specific molecular fingerprint are arrayed (Figure 4). This diagnostic test examining levels of the biomolecules within the COPD molecular fingerprint would transform the accuracy, reliability and reproducibility of COPD diagnosis and assessment. We discuss below the broad methodology of the chip design and analytical implementation that offers much promise with disease detection and management.
A schematic representation demonstrating how a COPD-specific SPR microarray chip could be employed. A small blood sample would be required, which would be separated into serum and cellular components using a microfluidic approach. Varying gene and protein expression would be monitored by changes in SPR enabling label free detection.
Target molecules
Initially the COPD specific microarray chip would be arrayed with antibodies, oligonucleotides and antigens as there is evidence of their ligands (proteins, RNA and antibodies respectively) being dysregulated in COPD [55, 195–197]. Whilst the level of complexity of a biological system is vast, incorporating multiple cellular, genetic and molecular components, current approaches to disease-specific pattern analysis focus on deciphering panels of only one molecular component i.e., protein or mRNA [50, 198, 199]. For a more comprehensive depiction of the disease state, however, simultaneous examination of both the mRNA and protein levels of a molecule is vital as evidence suggests that correlation between the two can be poor [200, 201]. In a study examining mRNA and protein expression in lung adenocarcinomas, only 21.4% of genes showed significant correlation with their corresponding protein [201]. Thus, both the mRNA and protein species of a molecule will be examined even if only one of these has been associated with disease. As the molecular fingerprint of COPD is further refined, the repertoire of detection would be adapted to allow for detection of single nucleotide polymorphisms (SNPs), microRNAs, peptides, enzymes/substrate interactions, small molecules (e.g. serotonin, vitamins, histamine), sugars or cell surface markers as appropriate.
Clinical sample type
Another important factor to consider is the source of clinical sample being examined. Samples traditionally examined in cases of respiratory disease include induced sputum, BAL, lung tissue and, more recently, exhaled breath condensate (EBC). All of these sample types could potentially be analysed for patterns of biomarkers, but they are hindered by their invasiveness, cost or high level of variability [202]. The systemic manifestations of many complex diseases, including COPD [11, 12], make analysis of body fluids an appealing option. In particular, the dynamic nature of blood means that it reflects the diverse physiological or pathological states of an individual. Coupled with its comparative ease of sampling, this makes the analysis of blood components the ultimate target for biomarker applications. Utilising blood samples would provide the opportunity to examine a full spectrum of molecular and cellular components within the disease-specific fingerprint including (but not exclusively) soluble proteins [50], cell types [203], cellular proteins/markers [204], autoantibodies [205], post-translational modifications [206] and circulating nucleic acids [207, 208]. The proposed use of whole blood as a sample would require steps for separation on the basis of size and the ability to lyse cells to extract intracellular components. This could be achieved by coupling a microfluidic system, such as that previously described [209], to the chip to allow in-situ separation of the blood sample into plasma and cellular components.
Despite the huge potential of blood samples in diagnostic tests, some major challenges with its implementation need to be overcome. Past investigations into plasma biomarkers have been hindered by the fact that the plasma proteome is dominated by several highly abundant proteins, which mask proteins of much lower abundance identified as contributing to disease states [210–212]. This is not a trivial problem even in cases in which highly selective molecular-recognition-based protein identification technologies, such as those which are antibody-based, are employed. It is also important to consider other factors that may affect serum protein levels including psychological stress, time of blood sample collection, time since last meal, or uncontrolled differences in specimen handling [213, 214]. Many of these limitations are beginning to be addressed [215, 216] increasing the feasibility of comprehensive diagnostic testing in plasma. To this end, preliminary studies examining patterns of biomolecules, including proteins and autoantibodies, have been undertaken with some success for diseases such as graft versus host disease [217], chronic pancreatitis [198], brain cancer [218], lung cancer [219, 220] and idiopathic pulmonary fibrosis (IPF) [221].
With regards to COPD, there is preliminary evidence that patterns of biomarkers in the peripheral compartment could be used to distinguish patients with COPD. Increased concentrations of TNF-α and IL-6 have been demonstrated in the serum of stable COPD patients [222]. Pinto-Plata et al used a protein microarray platform to identify 24 serum proteins that were up-regulated in COPD [50] whilst Shaker et al demonstrated that down regulation, as well as up-regulation, of plasma proteins was indicative of COPD [48]. Man et al took this one step further and demonstrated that ratios of blood biomarkers, in this case fibronectin and CRP, are significantly associated with all-cause mortality of COPD patients [52]. Whilst such studies should be considered a proof of principle rather than a comprehensive analysis of all possible biomolecules related to COPD, this data provides evidence that a systems biology approach to COPD diagnosis and evaluation is attainable within blood. Additionally, whilst forming a complex network of interaction in the lung, all the potential COPD biomarkers identified in Table 1 have been detected within blood (Figure 5), although this has not always been in the context of COPD. These molecules, combined with those identified by the aforementioned studies, could provide the basis of a prototype peripheral compartment COPD molecular fingerprint.
Defining, revising and analysing a molecular fingerprint
In addition to developing hardware with exquisite molecular sensitivity, the key to implementing advanced detection modalities is to include analytical protocols that are able to recognise complex biomolecular patterns made up of different molecular species and relate these to the disease condition under consideration e.g., COPD. Such analytical models now typically involve Bayesian inference approaches often starting with the hidden Markov model (HMM). This is essentially the simplest dynamic Bayesian network in which the system being studied is assumed to be a Markov process with unknown parameters. The challenge is to determine the hidden (i.e., disease) parameters from the observable molecular data so that the target condition of COPD can be identified. The Bayesian approach is particularly helpful with determination of the probability that any 'positive' result is actually a false positive. A systems biology approach to disease diagnosis strives to identify the presence of a molecular fingerprint of biomolecules that is not typically normal. Thus an observed biomolecular pattern from a suspected COPD patient is compared to a standardized 'healthy' pattern and diagnosed as having COPD or not. This approach is much more powerful than a diagnosis based on the presence of an altered concentration of a singular molecular marker e.g., PSA as it is less susceptible to the large variations in molecular marker concentration that naturally occur in any given population. The holistic measurement of a biomolecular pattern is more likely to reflect a disease condition than an individual molecular marker and, therefore, would augment the detection process. We are not alone in this vision, as others have also adopted this strategy as a way forward in molecular analysis. Alagaratnam et al are utilising Bayesian approaches to pursue muscular dystrophy diagnosis [223]. Similarly, the example we use above regarding PSA is also addressed using a systems analysis based on pattern-matching algorithms by groups in the US [224]. The problems with all these approaches however, are that they mostly rely on mass spectrometry for the molecular measurement and as such are expensive, require a significant investment in operator-skill and are less high-throughput than the SPR methodology we describe above. The latter point is extremely important if community screening is to be employed. Similarly, Bayesian approaches are not the only ways forward in mining the profile information. Other groups have discussed these approaches so we do not cover this in this review [225–227], but emphasise that it is the patterns of data that are important and not individual measurements. These analytical approaches are not just exclusive to the biomedical sciences as pattern analysis is central to much image analysis and recognition, such areas could well offer rich sources of analytical protocols.
Potential Benefits
Adopting an SPR-based systems biology approach to COPD diagnosis would provide several distinct benefits. The potential for vastly improved disease diagnosis and classification is evident. As described earlier, whilst the current method of COPD diagnosis, i.e. spirometry, provides an indication of airway obstruction, it is insufficient for accurate disease evaluation, classification and subtyping. Analysis of biomolecular patterns would provide details on the molecular and cellular basis underlying the onset of COPD in an individual facilitating highly accurate disease diagnosis and classification. It would also provide a means by which the health of a COPD patient could be efficiently monitored. Inclusion of multiple molecular species within the molecular fingerprint will provide far more information than that obtained by analysis of a single molecular species. Highlighting the stage at which expression levels of a molecule vary would provide a greater insight into the causes of disease onset, identify important pathways for further examination and help direct future treatment strategies. Having a greater understanding of the molecular profiles underlying COPD would pave the way for personalized medicine where drug treatments are tailored towards the causal factors of disease for each individual.
Early symptoms of COPD are chronic cough and sputum production, which are often ignored by the patients and physicians, as they are thought to be a normal consequence of smoking [228]. It is not until an individual experiences further airway obstruction that spirometric testing will be undertaken, by which time irreversible damage will have occurred. The longer such symptoms are ignored, the worse the decline in lung function will be. With early detection, however, it may be possible to slow the age-related decline in lung function [229]. Thus, it is necessary to find ways in which to diagnose COPD when it is at a stage that is treatable and when smoking cessation will have an effect on prognosis. An SPR-based systems biology approach to COPD diagnosis would allow regular examination of biomolecular patterns in individuals with a family history of disease or those who are exposed to disease risk factors. Monitoring such individuals should facilitate significant improvements in early disease detection allowing enhanced drug intervention and anti-smoking measures at a time when treatment will be more effective, improving prospects for life expectancy and quality.
Finally, the benefits of biomolecular patterns would be seen in the field of drug discovery and development. Adoption of this strategy could be used to circumvent some of the problems associated with phase III clinical trials during drug development. Currently the assessment of therapeutic efficacy in phase III COPD drug trials involves following a large number of patients, over a long period of time, in order to measure decline in FEV1. The finding of a disease specific profile that accurately reflects current disease activity would reduce the need for such long-term, expensive, clinical trials [230] by allowing assessment of the immediate impact of potential drug therapeutics on disease mechanisms prior to an improvement of outwardly detectable symptoms. Improved understanding of the cellular and molecular basis of COPD pathogenesis would also potentially provide new therapeutic targets.
Conclusion
Current methods for diagnosing COPD rely on spirometry combined with the use of questionnaires and other arbitrary measures for disease classification. Adopting a systems biology approach, whereby a disease defining molecular fingerprint is analysed, would increase the accuracy of disease diagnosis, aid earlier disease detection, allow for improved clarification of disease subtypes and allow automation for community screening.
References
Celli BR, MacNee W: Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J 2004,23(6):932–946.
Dennis RJ, Maldonado D, Norman S, Baena E, Castano H, Martinez G, Velez JR: Wood smoke exposure and risk for obstructive airways disease among women. Chest 1996,109(3 Suppl):55S-56S.
Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J: Developing COPD: a 25 year follow up study of the general population. Thorax 2006,61(11):935–939.
Brown C, Crombie I, Tunstall-Pedoe H: Failure of cigarette smoking to explain international differences in mortality from chronic obstructive pulmonary disease. J Epidemiol Community Health 1994,48(2):134–139.
Mannino DM, Buist AS: Global burden of COPD: risk factors, prevalence, and future trends. Lancet 2007,370(9589):765–773.
Jiang R, Paik DC, Hankinson JL, Barr RG: Cured meat consumption, lung function, and chronic obstructive pulmonary disease among United States adults. Am J Respir Crit Care Med 2007,175(8):798–804.
Barnes PJ: Small airways in COPD. N Engl J Med 2004,350(26):2635–2637.
Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease. American Thoracic Society Am J Respir Crit Care Med 1995,152(5 Pt 2):S77–121.
Mannino DM, Watt G, Hole D, Gillis C, Hart C, McConnachie A, Davey Smith G, Upton M, Hawthorne V, Sin DD, Man SF, Van Eeden S, Mapel DW, Vestbo J: The natural history of chronic obstructive pulmonary disease. Eur Respir J 2006,27(3):627–643.
Agusti A: Systemic effects of chronic obstructive pulmonary disease: what we know and what we don't know (but should). Proc Am Thorac Soc 2007,4(7):522–525.
Agusti A, Soriano JB: COPD as a Systemic Disease. Copd 2008,5(2):133–138.
Fabbri LM, Rabe KF: From COPD to chronic systemic inflammatory syndrome? Lancet 2007,370(9589):797–799.
Bernard S, LeBlanc P, Whittom F, Carrier G, Jobin J, Belleau R, Maltais F: Peripheral muscle weakness in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1998,158(2):629–634.
Rahman I, Morrison D, Donaldson K, MacNee W: Systemic oxidative stress in asthma, COPD, and smokers. Am J Respir Crit Care Med 1996,154(4 Pt 1):1055–1060.
Kamischke A, Kemper DE, Castel MA, Luthke M, Rolf C, Behre HM, Magnussen H, Nieschlag E: Testosterone levels in men with chronic obstructive pulmonary disease with or without glucocorticoid therapy. Eur Respir J 1998,11(1):41–45.
Gomez FP, Rodriguez-Roisin R: Global Initiative for Chronic Obstructive Lung Disease (GOLD) guidelines for chronic obstructive pulmonary disease. Curr Opin Pulm Med 2002,8(2):81–86.
Mahler DA, Weinberg DH, Wells CK, Feinstein AR: The measurement of dyspnea. Contents, interobserver agreement, and physiologic correlates of two new clinical indexes. Chest 1984,85(6):751–758.
Jones PW, Quirk FH, Baveystock CM, Littlejohns P: A self-complete measure of health status for chronic airflow limitation. The St. George's Respiratory Questionnaire. Am Rev Respir Dis 1992,145(6):1321–1327.
Alsaeedi A, Sin DD, McAlister FA: The effects of inhaled corticosteroids in chronic obstructive pulmonary disease: a systematic review of randomized placebo-controlled trials. Am J Med 2002,113(1):59–65.
O'Donnell DE, Lam M, Webb KA: Measurement of symptoms, lung hyperinflation, and endurance during exercise in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1998,158(5 Pt 1):1557–1565.
Celli BR, Cote CG, Marin JM, Casanova C, Montes de Oca M, Mendez RA, Pinto Plata V, Cabral HJ: The body-mass index, airflow obstruction, dyspnea, and exercise capacity index in chronic obstructive pulmonary disease. N Engl J Med 2004,350(10):1005–1012.
Ong KC, Earnest A, Lu SJ: A multidimensional grading system (BODE index) as predictor of hospitalization for COPD. Chest 2005,128(6):3810–3816.
Coultas DB, Mapel DW: Undiagnosed airflow obstruction: prevalence and implications. Curr Opin Pulm Med 2003,9(2):96–103.
Lundback B, Lindberg A, Lindstrom M, Ronmark E, Jonsson AC, Jonsson E, Larsson LG, Andersson S, Sandstrom T, Larsson K: Not 15 but 50% of smokers develop COPD?–Report from the Obstructive Lung Disease in Northern Sweden Studies. Respir Med 2003,97(2):115–122.
Rutschmann OT, Janssens JP, Vermeulen B, Sarasin FP: Knowledge of guidelines for the management of COPD: a survey of primary care physicians. Respir Med 2004,98(10):932–937.
Bolton CE, Ionescu AA, Edwards PH, Faulkner TA, Edwards SM, Shale DJ: Attaining a correct diagnosis of COPD in general practice. Respir Med 2005,99(4):493–500.
Jensen RL, Teeter JG, England RD, Howell HM, White HJ, Pickering EH, Crapo RO: Sources of long-term variability in measurements of lung function: implications for interpretation and clinical trial design. Chest 2007,132(2):396–402.
Macintyre N: Finding signals amidst the noise in pulmonary function testing. Chest 2007,132(2):367–368.
Hanania NA: The impact of inhaled corticosteroid and long-acting beta-agonist combination therapy on outcomes in COPD. Pulm Pharmacol Ther 2008,21(3):540–550.
Mauad T, Dolhnikoff M: Pathologic similarities and differences between asthma and chronic obstructive pulmonary disease. Curr Opin Pulm Med 2008,14(1):31–38.
Kalsheker N, Chappell S: The new genetics and chronic obstructive pulmonary disease. Copd 2008,5(4):257–264.
Demeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, Haley KJ, Sylvia JS, Sparrow D, Spira AE, Beane J, Pinto-Plata V, Speizer FE, Shapiro SD, Weiss ST, Silverman EK: The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006,78(2):253–264.
Hizawa N, Makita H, Nasuhara Y, Hasegawa M, Nagai K, Ito Y, Betsuyaku T, Konno S, Nishimura M: Functional single nucleotide polymorphisms of the CCL5 gene and nonemphysematous phenotype in COPD patients. Eur Respir J 2008,32(2):372–378.
Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, Province MA, Rao DC, Reilly JJ, Ginns LC, Speizer FE, Weiss ST: Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet 2002,70(5):1229–1239.
Chappell S, Daly L, Morgan K, Guetta Baranes T, Roca J, Rabinovich R, Millar A, Donnelly SC, Keatings V, MacNee W, Stolk J, Hiemstra P, Miniati M, Monti S, O'Connor CM, Kalsheker N: Cryptic haplotypes of SERPINA1 confer susceptibility to chronic obstructive pulmonary disease. Hum Mutat 2006,27(1):103–109.
Hersh CP, Hansel NN, Barnes KC, Lomas DA, Pillai SG, Coxson HO, Mathias RA, Rafaels NM, Wise RA, Connett JE, Klanderman BJ, Jacobson FL, Gill R, Litonjua AA, Sparrow D, Reilly JJ, Silverman EK: Transforming Growth Factor Beta Receptor-3 is Associated with Pulmonary Emphysema. Am J Respir Cell Mol Biol 2009, in press.
Pierrou S, Broberg P, O'Donnell RA, Pawlowski K, Virtala R, Lindqvist E, Richter A, Wilson SJ, Angco G, Moller S, Bergstrand H, Koopmann W, Wieslander E, Stromstedt PE, Holgate ST, Davies DE, Lund J, Djukanovic R: Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007,175(6):577–586.
Wood AM, Stockley RA: The genetics of chronic obstructive pulmonary disease. Respir Res 2006, 7:130.
Wang IM, Stepaniants S, Boie Y, Mortimer JR, Kennedy B, Elliott M, Hayashi S, Loy L, Coulter S, Cervino S, Harris J, Thornton M, Raubertas R, Roberts C, Hogg JC, Crackower M, O'Neill G, Pare PD: Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med 2008,177(4):402–411.
Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, Pinsky DJ, Watkins SC, Pilewski JM, Sciurba FC, Peters DG, Hogg JC, Choi AM: Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA 2004,101(41):14895–14900.
Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF: Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol 2004,31(6):595–600.
Betsuyaku T, Fuke S, Nasuhara Y, Morikawa T, Kondo S, Nishimura M: Diverse expression of antioxidants and inflammatory chemokines in terminal bronchiolar epithelium in chronic obstructive pulmonary disease. Proc Am Thorac Soc 2006,3(6):471–472.
Bhattacharya S, Srisuma S, Demeo DL, Reilly JJ, Bueno R, Silverman EK, Mariani TJ: Microarray data-based prioritization of chronic obstructive pulmonary disease susceptibility genes. Proc Am Thorac Soc 2006,3(6):472.
Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS: Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol 2004,31(6):601–610.
Bhattacharya S, Srisuma S, Demeo DL, Shapiro SD, Bueno R, Silverman EK, Reilly JJ, Mariani TJ: Molecular biomarkers for quantitative and discrete COPD phenotypes. Am J Respir Cell Mol Biol 2009,40(3):359–367.
Zhang W, Yan SD, Zhu A, Zou YS, Williams M, Godman GC, Thomashow BM, Ginsburg ME, Stern DM, Yan SF: Expression of Egr-1 in late stage emphysema. Am J Pathol 2000,157(4):1311–1320.
Gessner C, Scheibe R, Wotzel M, Hammerschmidt S, Kuhn H, Engelmann L, Hoheisel G, Gillissen A, Sack U, Wirtz H: Exhaled breath condensate cytokine patterns in chronic obstructive pulmonary disease. Respir Med 2005,99(10):1229–1240.
Shaker SB, von Wachenfeldt KA, Larsson S, Mile I, Persdotter S, Dahlbäck M, Broberg P, Stoel B, Bach KS, Hestad M, Fehniger TE, Dirksen A: Identification of patients with chronic obstructive pulmonary disease (COPD) by measurement of plasma biomarkers. Clin Resp J 2008,2(1):17–25.
Plymoth A, Lofdahl CG, Ekberg-Jansson A, Dahlback M, Broberg P, Foster M, Fehniger TE, Marko-Varga G: Protein expression patterns associated with progression of chronic obstructive pulmonary disease in bronchoalveolar lavage of smokers. Clin Chem 2007,53(4):636–644.
Pinto-Plata V, Toso J, Lee K, Park D, Bilello J, Mullerova H, De Souza MM, Vessey R, Celli B: Profiling serum biomarkers in patients with COPD: associations with clinical parameters. Thorax 2007,62(7):595–601.
Braido F, Riccio AM, Guerra L, Gamalero C, Zolezzi A, Tarantini F, De Giovanni B, Folli C, Descalzi D, Canonica GW: Clara cell 16 protein in COPD sputum: a marker of small airways damage? Respir Med 2007,101(10):2119–2124.
Man SF, Xing L, Connett JE, Anthonisen NR, Wise RA, Tashkin DP, Zhang X, Vessey R, Walker TG, Celli BR, Sin DD: Circulating fibronectin to C-reactive protein ratio and mortality: a biomarker in COPD? Eur Respir J 2008,32(6):1451–1457.
Bacakoglu F, Atasever A, Ozhan MH, Gurgun C, Ozkilic H, Guzelant A: Plasma and bronchoalveolar lavage fluid levels of endothelin-1 in patients with chronic obstructive pulmonary disease and pulmonary hypertension. Respiration 2003,70(6):594–599.
Panzner P, Lafitte JJ, Tsicopoulos A, Hamid Q, Tulic MK: Marked up-regulation of T lymphocytes and expression of interleukin-9 in bronchial biopsies from patients With chronic bronchitis with obstruction. Chest 2003,124(5):1909–1915.
Kent L, Smyth L, Clayton C, Scott L, Cook T, Stephens R, Fox S, Hext P, Farrow S, Singh D: Cigarette smoke extract induced cytokine and chemokine gene expression changes in COPD macrophages. Cytokine 2008,42(2):205–216.
Barcelo B, Pons J, Fuster A, Sauleda J, Noguera A, Ferrer JM, Agusti AG: Intracellular cytokine profile of T lymphocytes in patients with chronic obstructive pulmonary disease. Clin Exp Immunol 2006,145(3):474–479.
Bucchioni E, Kharitonov SA, Allegra L, Barnes PJ: High levels of interleukin-6 in the exhaled breath condensate of patients with COPD. Respir Med 2003,97(12):1299–1302.
Hackett TL, Holloway R, Holgate ST, Warner JA: Dynamics of pro-inflammatory and anti-inflammatory cytokine release during acute inflammation in Chronic Obstructive Pulmonary Disease: An ex vivo study. Respir Res 2008,9(1):47.
Tsoumakidou M, Demedts IK, Brusselle GG, Jeffery PK: Dendritic cells in chronic obstructive pulmonary disease: new players in an old game. Am J Respir Crit Care Med 2008,177(11):1180–1186.
Di Stefano A, Caramori G, Capelli A, Gnemmi I, Ricciardolo FL, Oates T, Donner CF, Chung KF, Barnes PJ, Adcock IM: STAT4 activation in smokers and patients with chronic obstructive pulmonary disease. Eur Respir J 2004,24(1):78–85.
Lee JS, Rosengart MR, Kondragunta V, Zhang Y, McMurray J, Branch RA, Choi AM, Sciurba FC: Inverse association of plasma IL-13 and inflammatory chemokines with lung function impairment in stable COPD: a cross-sectional cohort study. Respir Res 2007, 8:64.
Hegab AE, Sakamoto T, Saitoh W, Massoud HH, Massoud HM, Hassanein KM, Sekizawa K: Polymorphisms of IL4, IL13, and ADRB2 genes in COPD. Chest 2004,126(6):1832–1839.
Imaoka H, Hoshino T, Takei S, Kinoshita T, Okamoto M, Kawayama T, Kato S, Iwasaki H, Watanabe K, Aizawa H: Interleukin-18 production and pulmonary function in COPD. Eur Respir J 2008,31(2):287–297.
Saetta M, Mariani M, Panina-Bordignon P, Turato G, Buonsanti C, Baraldo S, Bellettato CM, Papi A, Corbetta L, Zuin R, Sinigaglia F, Fabbri LM: Increased expression of the chemokine receptor CXCR3 and its ligand CXCL10 in peripheral airways of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2002,165(10):1404–1409.
Gingo MR, Silveira LJ, Miller YE, Friedlander AL, Cosgrove GP, Chan ED, Maier LA, Bowler RP: Tumour necrosis factor gene polymorphisms are associated with COPD. Eur Respir J 2008,31(5):1005–1012.
Lee SH, Goswami S, Grudo A, Song LZ, Bandi V, Goodnight-White S, Green L, Hacken-Bitar J, Huh J, Bakaeen F, Coxson HO, Cogswell S, Storness-Bliss C, Corry DB, Kheradmand F: Antielastin autoimmunity in tobacco smoking-induced emphysema. Nat Med 2007,13(5):567–569.
Baraldo S, Bazzan E, Zanin ME, Turato G, Garbisa S, Maestrelli P, Papi A, Miniati M, Fabbri LM, Zuin R, Saetta M: Matrix metalloproteinase-2 protein in lung periphery is related to COPD progression. Chest 2007,132(6):1733–1740.
Molet S, Belleguic C, Lena H, Germain N, Bertrand CP, Shapiro SD, Planquois JM, Delaval P, Lagente V: Increase in macrophage elastase (MMP-12) in lungs from patients with chronic obstructive pulmonary disease. Inflamm Res 2005,54(1):31–36.
Andelid K, Bake B, Rak S, Linden A, Rosengren A, Ekberg-Jansson A: Myeloperoxidase as a marker of increasing systemic inflammation in smokers without severe airway symptoms. Respir Med 2007,101(5):888–895.
Starosta V, Ratjen F, Rietschel E, Paul K, Griese M: Anti-inflammatory cytokines in cystic fibrosis lung disease. Eur Respir J 2006,28(3):581–587.
Chen J, Lam S, Pilon A, McWilliams A, Macaulay C, Szabo E: Higher levels of the anti-inflammatory protein CC10 are associated with improvement in bronchial dysplasia and sputum cytometric assessment in individuals at high risk for lung cancer. Clin Cancer Res 2008,14(5):1590–1597.
Lee JG, Cho BC, Bae MK, Lee CY, Park IK, Kim DJ, Ahn SV, Chung KY: Preoperative C-reactive protein levels are associated with tumor size and lymphovascular invasion in resected non-small cell lung cancer. Lung Cancer 2009,63(1):106–110.
Ueno T, Kataoka M, Hirano A, Iio K, Tanimoto Y, Kanehiro A, Okada C, Soda R, Takahashi K, Tanimoto M: Inflammatory markers in exhaled breath condensate from patients with asthma. Respirology 2008,13(5):654–663.
Fagan KA, McMurtry IF, Rodman DM: Role of endothelin-1 in lung disease. Respir Res 2001,2(2):90–101.
Hill TA, Lightman S, Pantelidis P, Abdallah A, Spagnolo P, du Bois RM: Intracellular cytokine profiles and T cell activation in pulmonary sarcoidosis. Cytokine 2008,42(3):289–292.
Fulton RB, Olson MR, Varga SM: Regulation of cytokine production by virus-specific CD8 T cells in the lungs. J Virol 2008,82(16):7799–7811.
Nahm DH, Lee YE, Yim EJ, Park HS, Yim H, Kang Y, Kim JK: Identification of cytokeratin 18 as a bronchial epithelial autoantigen associated with nonallergic asthma. Am J Respir Crit Care Med 2002,165(11):1536–1539.
Maniwa K, Ogushi F, Tani K, Ohmoto Y, Muraguchi M, Sone S: Increased incidence of autoantibodies to interleukin-1a in rheumatoid arthritis with interstitial lung disease. Respirology 2000,5(4):315–320.
Eastgate JA, Symons JA, Wood NC, Grinlinton FM, di Giovine FS, Duff GW: Correlation of plasma interleukin 1 levels with disease activity in rheumatoid arthritis. Lancet 1988,2(8613):706–709.
Al-Shukaili A, Al-Kaabi J, Hassan B: A comparative study of interleukin-1beta production and p2 × 7 expression after ATP stimulation by peripheral blood mononuclear cells isolated from rheumatoid arthritis patients and normal healthy controls. Inflammation 2008,31(2):84–90.
Turzanski J, Grundy M, Russell NH, Pallis M: Interleukin-1beta maintains an apoptosis-resistant phenotype in the blast cells of acute myeloid leukaemia via multiple pathways. Leukemia 2004,18(10):1662–1670.
Loza MJ, Chang BL: Association between Q551R IL4R genetic variants and atopic asthma risk demonstrated by meta-analysis. J Allergy Clin Immunol 2007,120(3):578–585.
Chatila TA: Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med 2004,10(10):493–499.
Yeh HH, Lai WW, Chen HH, Liu HS, Su WC: Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene 2006,25(31):4300–4309.
Takizawa H, Satoh M, Okazaki H, Matsuzaki G, Suzuki N, Ishii A, Suko M, Okudaira H, Morita Y, Ito K: Increased IL-6 and IL-8 in bronchoalveolar lavage fluids (BALF) from patients with sarcoidosis: correlation with the clinical parameters. Clin Exp Immunol 1997,107(1):175–181.
Nocker RE, Schoonbrood DF, Graaf EA, Hack CE, Lutter R, Jansen HM, Out TA: Interleukin-8 in airway inflammation in patients with asthma and chronic obstructive pulmonary disease. Int Arch Allergy Immunol 1996,109(2):183–191.
Henriquet C, Gougat C, Combes A, Lazennec G, Mathieu M: Differential regulation of RANTES and IL-8 expression in lung adenocarcinoma cells. Lung Cancer 2007,56(2):167–174.
Fujimori Y, Kataoka M, Tada S, Takehara H, Matsuo K, Miyake T, Okahara M, Yamadori I, Tanimoto M: The role of interleukin-8 in interstitial pneumonia. Respirology 2003,8(1):33–40.
Ogden CA, Pound JD, Batth BK, Owens S, Johannessen I, Wood K, Gregory CD: Enhanced apoptotic cell clearance capacity and B cell survival factor production by IL-10-activated macrophages: implications for Burkitt's lymphoma. J Immunol 2005,174(5):3015–3023.
Larche M: Regulatory T cells in allergy and asthma. Chest 2007,132(3):1007–1014.
Scumpia PO, Moldawer LL: Biology of interleukin-10 and its regulatory roles in sepsis syndromes. Crit Care Med 2005,33(12 Suppl):S468–471.
Peluso I, Pallone F, Monteleone G: Interleukin-12 and Th1 immune response in Crohn's disease: pathogenetic relevance and therapeutic implication. World J Gastroenterol 2006,12(35):5606–5610.
Huang X, Hua J, Shen N, Chen S: Dysregulated expression of interleukin-23 and interleukin-12 subunits in systemic lupus erythematosus patients. Mod Rheumatol 2007,17(3):220–223.
Saha SK, Berry MA, Parker D, Siddiqui S, Morgan A, May R, Monk P, Bradding P, Wardlaw AJ, Pavord ID, Brightling CE: Increased sputum and bronchial biopsy IL-13 expression in severe asthma. J Allergy Clin Immunol 2008,121(3):685–691.
Wills-Karp M: Interleukin-13 in asthma pathogenesis. Immunol Rev 2004, 202:175–190.
Pawlik A, Kaminski M, Kusnierczyk P, Kurzawski M, Dziedziejko V, Adamska M, Safranow K, Gawronska-Szklarz B: Interleukin-18 promoter polymorphism in patients with atopic asthma. Tissue Antigens 2007,70(4):314–318.
Kieszko R, Krawczyk P, Jankowska O, Chocholska S, Krol A, Milanowski J: The clinical significance of interleukin 18 assessment in sarcoidosis patients. Respir Med 2007,101(4):722–728.
Nureki S, Miyazaki E, Ando M, Ueno T, Fukami T, Kumamoto T, Sugisaki K, Tsuda T: Circulating levels of both Th1 and Th2 chemokines are elevated in patients with sarcoidosis. Respir Med 2008,102(2):239–247.
Wark PA, Bucchieri F, Johnston SL, Gibson PG, Hamilton L, Mimica J, Zummo G, Holgate ST, Attia J, Thakkinstian A, Davies DE: IFN-gamma-induced protein 10 is a novel biomarker of rhinovirus-induced asthma exacerbations. J Allergy Clin Immunol 2007,120(3):586–593.
Tang NL, Chan PK, Wong CK, To KF, Wu AK, Sung YM, Hui DS, Sung JJ, Lam CW: Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin Chem 2005,51(12):2333–2340.
Okamoto M, Kawabe T, Iwasaki Y, Hara T, Hashimoto N, Imaizumi K, Hasegawa Y, Shimokata K: Evaluation of interferon-gamma, interferon-gamma-inducing cytokines, and interferon-gamma-inducible chemokines in tuberculous pleural effusions. J Lab Clin Med 2005,145(2):88–93.
Iniesta P, Moran A, De Juan C, Gomez A, Hernando F, Garcia-Aranda C, Frias C, Diaz-Lopez A, Rodriguez-Jimenez FJ, Balibrea JL, Benito M: Biological and clinical significance of MMP-2, MMP-9, TIMP-1 and TIMP-2 in non-small cell lung cancer. Oncol Rep 2007,17(1):217–223.
Kanoh Y, Ohara T, Tadano T, Kanoh M, Akahoshi T: Changes to N-linked oligosaccharide chains of human serum immunoglobulin G and matrix metalloproteinase-2 with cancer progression. Anticancer Res 2008,28(2A):715–720.
Henderson N, Markwick LJ, Elshaw SR, Freyer AM, Knox AJ, Johnson SR: Collagen I and thrombin activate MMP-2 by MMP-14-dependent and -independent pathways: implications for airway smooth muscle migration. Am J Physiol Lung Cell Mol Physiol 2007,292(4):L1030–1038.
Matute-Bello G, Wurfel MM, Lee JS, Park DR, Frevert CW, Madtes DK, Shapiro SD, Martin TR: Essential role of MMP-12 in Fas-induced lung fibrosis. Am J Respir Cell Mol Biol 2007,37(2):210–221.
Nenan S, Boichot E, Lagente V, Bertrand CP: Macrophage elastase (MMP-12): a pro-inflammatory mediator? Mem Inst Oswaldo Cruz 2005,100(Suppl 1):167–172.
Hofmann HS, Hansen G, Richter G, Taege C, Simm A, Silber RE, Burdach S: Matrix metalloproteinase-12 expression correlates with local recurrence and metastatic disease in non-small cell lung cancer patients. Clin Cancer Res 2005,11(3):1086–1092.
Taioli E, Benhamou S, Bouchardy C, Cascorbi I, Cajas-Salazar N, Dally H, Fong KM, Larsen JE, Le Marchand L, London SJ, Risch A, Spitz MR, Stucker I, Weinshenker B, Wu X, Yang P: Myeloperoxidase G-463A polymorphism and lung cancer: a HuGE genetic susceptibility to environmental carcinogens pooled analysis. Genet Med 2007,9(2):67–73.
Reynolds WF, Sermet-Gaudelus I, Gausson V, Feuillet MN, Bonnefont JP, Lenoir G, Descamps-Latscha B, Witko-Sarsat V: Myeloperoxidase promoter polymorphism -463G is associated with more severe clinical expression of cystic fibrosis pulmonary disease. Mediators Inflamm 2006,2006(2):36735.
Kodama T, Yukioka H, Kato T, Kato N, Hato F, Kitagawa S: Neutrophil elastase as a predicting factor for development of acute lung injury. Intern Med 2007,46(11):699–704.
Yang P, Bamlet WR, Sun Z, Ebbert JO, Aubry MC, Krowka MJ, Taylor WR, Marks RS, Deschamps C, Swensen SJ, Wieben ED, Cunningham JM, Melton LJ, de Andrade M: Alpha1-antitrypsin and neutrophil elastase imbalance and lung cancer risk. Chest 2005,128(1):445–452.
Elizur A, Cannon CL, Ferkol TW: Airway inflammation in cystic fibrosis. Chest 2008,133(2):489–495.
Mori S, Gibson G, McTiernan CF: Differential expression of MMPs and TIMPs in moderate and severe heart failure in a transgenic model. J Card Fail 2006,12(4):314–325.
Higashimoto Y, Yamagata Y, Taya S, Iwata T, Okada M, Ishiguchi T, Sato H, Itoh H: Systemic inflammation in chronic obstructive pulmonary disease and asthma: Similarities and differences. Respirology 2008,13(1):128–133.
Nicol MQ, Mathys JM, Pereira A, Ollington K, Ieong MH, Skolnik PR: Human immunodeficiency virus infection alters tumor necrosis factor alpha production via Toll-like receptor-dependent pathways in alveolar macrophages and U1 cells. J Virol 2008,82(16):7790–7798.
Hanania NA: Targeting airway inflammation in asthma: current and future therapies. Chest 2008,133(4):989–998.
Anderson NL, Anderson NG: The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 2002,1(11):845–867.
Balbi B, Pignatti P, Corradi M, Baiardi P, Bianchi L, Brunetti G, Radaeli A, Moscato G, Mutti A, Spanevello A, Malerba M: Bronchoalveolar lavage, sputum and exhaled clinically relevant inflammatory markers: values in healthy adults. Eur Respir J 2007,30(4):769–781.
Crawford ED, Abrahamsson PA: PSA-based Screening for Prostate Cancer: How Does It Compare with Other Cancer Screening Tests? Eur Urol 2008,54(2):262–273.
Yarden Y: Biology of HER2 and its importance in breast cancer. Oncology 2001,61(Suppl 2):1–13.
Kirschner MW: The meaning of systems biology. Cell 2005,121(4):503–504.
Aderem A: Systems biology: its practice and challenges. Cell 2005,121(4):511–513.
Lucas P: Bayesian analysis, pattern analysis, and data mining in health care. Curr Opin Crit Care 2004,10(5):399–403.
Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, Friedman LF, Galasko DR, Jutel M, Karydas A, Kaye JA, Leszek J, Miller BL, Minthon L, Quinn JF, Rabinovici GD, Robinson WH, Sabbagh MN, So YT, Sparks DL, Tabaton M, Tinklenberg J, Yesavage JA, Tibshirani R, Wyss-Coray T: Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med 2007,13(11):1359–1362.
Knowlton N, Jiang K, Frank MB, Aggarwal A, Wallace C, McKee R, Chaser B, Tung C, Smith L, Chen Y, Osban J, O'Neil K, Centola M, McGhee JL, Jarvis JN: The meaning of clinical remission in polyarticular juvenile idiopathic arthritis: Gene expression profiling in peripheral blood mononuclear cells identifies distinct disease states. Arthritis Rheum 2009,60(3):892–900.
Reckow S, Gormanns P, Holsboer F, Turck CW: Psychiatric disorders biomarker identification: from proteomics to systems biology. Pharmacopsychiatry 2008,41(Suppl 1):S70–77.
Hu S, Zhou M, Jiang J, Wang J, Elashoff D, Gorr S, Michie SA, Spijkervet FK, Bootsma H, Kallenberg CG, Vissink A, Horvath S, Wong DT: Systems biology analysis of Sjogren's syndrome and mucosa-associated lymphoid tissue lymphoma in parotid glands. Arthritis Rheum 2009,60(1):81–92.
Sieberts SK, Schadt EE: Moving toward a system genetics view of disease. Mamm Genome 2007,18(6–7):389–401.
Greef J, Stroobant P, Heijden R: The role of analytical sciences in medical systems biology. Curr Opin Chem Biol 2004,8(5):559–565.
Hanash SM, Pitteri SJ, Faca VM: Mining the plasma proteome for cancer biomarkers. Nature 2008,452(7187):571–579.
Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Stattin P, Liu W, Kim JW, Duggan D, Carpten J, Isaacs W, Gronberg H, Xu J, Chang BL: Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res 2009,69(1):10–15.
Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A: MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther 2009.,8(4):
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY: MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna 2008,14(11):2348–2360.
Lebrilla CB, An HJ: The prospects of glycan biomarkers for the diagnosis of diseases. Mol Biosyst 2009,5(1):17–20.
Lim YP: Mining the tumor phosphoproteome for cancer markers. Clin Cancer Res 2005,11(9):3163–3169.
Elshal MF, McCoy JP: Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods 2006,38(4):317–323.
Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA: Mass Spectrometry Based Targeted Protein Quantification: Methods and Applications. J Proteome Res 2009,8(2):787–797.
Feng X, Liu X, Luo Q, Liu BF: Mass spectrometry in systems biology: an overview. Mass Spectrom Rev 2008,27(6):635–660.
Smith JC, Figeys D: Proteomics technology in systems biology. Mol Biosyst 2006,2(8):364–370.
Sheta EA, Appel SH, Goldknopf IL: 2D gel blood serum biomarkers reveal differential clinical proteomics of the neurodegenerative diseases. Expert Rev Proteomics 2006,3(1):45–62.
Ramachandran N, Larson DN, Stark PR, Hainsworth E, LaBaer J: Emerging tools for real-time label-free detection of interactions on functional protein microarrays. Febs J 2005,272(21):5412–5425.
Yeo WS, Min DH, Hsieh RW, Greene GL, Mrksich M: Label-free detection of protein-protein interactions on biochips. Angew Chem Int Ed Engl 2005,44(34):5480–5483.
Cooper MA: Label-free screening of bio-molecular interactions. Anal Bioanal Chem 2003,377(5):834–842.
Mullett WM, Lai EP, Yeung JM: Surface plasmon resonance-based immunoassays. Methods 2000,22(1):77–91.
Alonso-Lomillo MA, Yardimci C, Dominguez-Renedo O, Arcos-Martinez MJ: CYP450 2B4 covalently attached to carbon and gold screen printed electrodes by diazonium salt and thiols monolayers. Anal Chim Acta 2009,633(1):51–56.
Solanki PR, Prabhakar N, Pandey MK, Malhotra BD: Self-assembled monolayer for toxicant detection using nucleic acid sensor based on surface plasmon resonance technique. Biomed Microdevices 2008,10(5):757–767.
Kim HC, Lee SK, Jeon WB, Lyu HK, Lee SW, Jeong SW: Detection of C-reactive protein on a functional poly(thiophene) self-assembled monolayer using surface plasmon resonance. Ultramicroscopy 2008,108(10):1379–1383.
Zhang Y, Luo S, Tang Y, Yu L, Hou KY, Cheng JP, Zeng X, Wang PG: Carbohydrate-protein interactions by "clicked" carbohydrate self-assembled monolayers. Anal Chem 2006,78(6):2001–2008.
Liljeblad M, Lundblad A, Pahlsson P: Analysis of agalacto-IgG in rheumatoid arthritis using surface plasmon resonance. Glycoconj J 2000,17(5):323–329.
Ito M, Nakamura F, Baba A, Tamada K, Ushijima H, Lau KHA, Manna A, Knoll W: Enhancement of surface plasmon resonance signals by gold nanoparticles on high-density DNA microarrays. J Phys Chem C 2002,111(31):11653–11662.
Ohno O, Ikeda Y, Sawa R, Igarashi M, Kinoshita N, Suzuki Y, Miyake K, Umezawa K: Isolation of heptadepsin, a novel bacterial cyclic depsipeptide that inhibits lipopolysaccharide activity. Chem Biol 2004,11(8):1059–1070.
Alam SM, Paleos CA, Liao HX, Scearce R, Robinson J, Haynes BF: An inducible HIV type 1 gp41 HR-2 peptide-binding site on HIV type 1 envelope gp120. AIDS Res Hum Retroviruses 2004,20(8):836–845.
Rodius S, Chaloin O, Moes M, Schaffner-Reckinger E, Landrieu I, Lippens G, Lin M, Zhang J, Kieffer N: The talin rod IBS2 alpha-helix interacts with the beta3 integrin cytoplasmic tail membrane-proximal helix by establishing charge complementary salt bridges. J Biol Chem 2008,283(35):24212–24223.
Coombe DR, Stevenson SM, Kinnear BF, Gandhi NS, Mancera RL, Osmond RI, Kett WC: Platelet endothelial cell adhesion molecule 1 (PECAM-1) and its interactions with glycosaminoglycans: 2. Biochemical analyses. Biochemistry 2008,47(17):4863–4875.
Tabata S, Kuroki K, Wang J, Kajikawa M, Shiratori I, Kohda D, Arase H, Maenaka K: Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J Biol Chem 2008,283(14):8893–8901.
Davis PM, Abraham R, Xu L, Nadler SG, Suchard SJ: Abatacept binds to the Fc receptor CD64 but does not mediate complement-dependent cytotoxicity or antibody-dependent cellular cytotoxicity. J Rheumatol 2007,34(11):2204–2210.
Hassan MI, Kumar V, Somvanshi RK, Dey S, Singh TP, Yadav S: Structure-guided design of peptidic ligand for human prostate specific antigen. J Pept Sci 2007,13(12):849–855.
Steckbeck JD, Grieser HJ, Sturgeon T, Taber R, Chow A, Bruno J, Murphy-Corb M, Montelaro RC, Cole KS: Dynamic evolution of antibody populations in a rhesus macaque infected with attenuated simian immunodeficiency virus identified by surface plasmon resonance. J Med Primatol 2006,35(4–5):248–260.
Liu R, Yuan B, Emadi S, Zameer A, Schulz P, McAllister C, Lyubchenko Y, Goud G, Sierks MR: Single chain variable fragments against beta-amyloid (Abeta) can inhibit Abeta aggregation and prevent abeta-induced neurotoxicity. Biochemistry 2004,43(22):6959–6967.
Miao B, Li J, Fu X, Ding J, Geng M: T-cell receptor (TCR)/CD3 is involved in sulfated polymannuroguluronate (SPMG)-induced T lymphocyte activation. Int Immunopharmacol 2005,5(7–8):1171–1182.
Huber W: A new strategy for improved secondary screening and lead optimization using high-resolution SPR characterization of compound-target interactions. J Mol Recognit 2005,18(4):273–281.
Lam SN, Acharya P, Wyatt R, Kwong PD, Bewley CA: Tyrosine-sulfate isosteres of CCR5 N-terminus as tools for studying HIV-1 entry. Bioorg Med Chem 2008,16(23):10113–10120.
Perez M, Cuadros R, Benitez MJ, Jimenez JS: Interaction of Alzheimer's disease amyloid beta peptide fragment 25–35 with tau protein, and with a tau peptide containing the microtubule binding domain. J Alzheimers Dis 2004,6(5):461–467.
Hasegawa K, Ono K, Yamada M, Naiki H: Kinetic modeling and determination of reaction constants of Alzheimer's beta-amyloid fibril extension and dissociation using surface plasmon resonance. Biochemistry 2002,41(46):13489–13498.
Sourial S, Nilsson C, Warnmark A, Achour A, Harris RA: Deletion of the V1/V2 region does not increase the accessibility of the V3 region of recombinant gp125. Curr HIV Res 2006,4(2):229–237.
Lee HJ, Nedelkov D, Corn RM: Surface plasmon resonance imaging measurements of antibody arrays for the multiplexed detection of low molecular weight protein biomarkers. Anal Chem 2006,78(18):6504–6510.
Yuk JS, Ha KS: Proteomic applications of surface plasmon resonance biosensors: analysis of protein arrays. Exp Mol Med 2005,37(1):1–10.
Ladd J, Taylor AD, Piliarik M, Homola J, Jiang S: Label-free detection of cancer biomarker candidates using surface plasmon resonance imaging. Anal Bioanal Chem 2009,393(4):1157–1163.
Hu WP, Hsu HY, Chiou A, Tseng KY, Lin HY, Chang GL, Chen SJ: Immunodetection of pentamer and modified C-reactive protein using surface plasmon resonance biosensing. Biosens Bioelectron 2006,21(8):1631–1637.
Masson JF, Obando L, Beaudoin S, Booksh K: Sensitive and real-time fiber-optic-based surface plasmon resonance sensors for myoglobin and cardiac troponin I. Talanta 2004,62(5):865–870.
Besselink GA, Kooyman RP, van Os PJ, Engbers GH, Schasfoort RB: Signal amplification on planar and gel-type sensor surfaces in surface plasmon resonance-based detection of prostate-specific antigen. Anal Biochem 2004,333(1):165–173.
Saerens D, Frederix F, Reekmans G, Conrath K, Jans K, Brys L, Huang L, Bosmans E, Maes G, Borghs G, Muyldermans S: Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal Chem 2005,77(23):7547–7555.
de Boer AR, Hokke CH, Deelder AM, Wuhrer M: Serum antibody screening by surface plasmon resonance using a natural glycan microarray. Glycoconj J 2008,25(1):75–84.
Feriotto G, Ferlini A, Ravani A, Calzolari E, Mischiati C, Bianchi N, Gambari R: Biosensor technology for real-time detection of the cystic fibrosis W1282X mutation in CFTR. Hum Mutat 2001,18(1):70–81.
Suda Y, Arano A, Fukui Y, Koshida S, Wakao M, Nishimura T, Kusumoto S, Sobel M: Immobilization and clustering of structurally defined oligosaccharides for sugar chips: an improved method for surface plasmon resonance analysis of protein-carbohydrate interactions. Bioconjug Chem 2006,17(5):1125–1135.
Blow N: Glycobiology: A spoonful of sugar. Nature 2009,457(7229):617–620.
Klenkar G, Liedberg B: A microarray chip for label-free detection of narcotics. Anal Bioanal Chem 2008,391(5):1679–1688.
Bailly-Chouriberry L, Chu-Van E, Pinel G, Garcia P, Popot MA, Andre-Fontaine G, Bonnaire Y, Le Bizec B: Detection of secondary biomarker of met-eGH as a strategy to screen for somatotropin misuse in horseracing. Analyst 2008,133(2):270–276.
Lung FD, Chen HY, Lin HT: Monitoring bone loss using ELISA and surface plasmon resonance (SPR) technology. Protein Pept Lett 2003,10(3):313–319.
Lung FD, Chen CH, Liou CC, Chen HY: Surface plasmon resonance detection of interactions between peptide fragments of N-telopeptide and its monoclonal antibodies. J Pept Res 2004,63(4):365–370.
Fu E, Chinowsky T, Nelson K, Johnston K, Edwards T, Helton K, Grow M, Miller JW, Yager P: SPR imaging-based salivary diagnostics system for the detection of small molecule analytes. Ann N Y Acad Sci 2007, 1098:335–344.
Fang S, Lee HJ, Wark AW, Corn RM: Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J Am Chem Soc 2006,128(43):14044–14046.
Battaglia TM, Masson JF, Sierks MR, Beaudoin SP, Rogers J, Foster KN, Holloway GA, Booksh KS: Quantification of cytokines involved in wound healing using surface plasmon resonance. Anal Chem 2005,77(21):7016–7023.
Lee SJ, Youn BS, Park JW, Niazi JH, Kim YS, Gu MB: ssDNA aptamer-based surface plasmon resonance biosensor for the detection of retinol binding protein 4 for the early diagnosis of type 2 diabetes. Anal Chem 2008,80(8):2867–2873.
Teramura Y, Iwata H: Label-free immunosensing for alpha-fetoprotein in human plasma using surface plasmon resonance. Anal Biochem 2007,365(2):201–207.
Masson JF, Battaglia TM, Khairallah P, Beaudoin S, Booksh KS: Quantitative measurement of cardiac markers in undiluted serum. Anal Chem 2007,79(2):612–619.
Kurita R, Yokota Y, Sato Y, Mizutani F, Niwa O: On-chip enzyme immunoassay of a cardiac marker using a microfluidic device combined with a portable surface plasmon resonance system. Anal Chem 2006,78(15):5525–5531.
Nagel T, Gajovic-Eichelmann N, Tobisch S, Schulte-Spechtel U, Bier FF: Serodiagnosis of Lyme borreliosis infection using surface plasmon resonance. Clin Chim Acta 2008,394(1–2):110–113.
Dutra RF, Mendes RK, Lins da Silva V, Kubota LT: Surface plasmon resonance immunosensor for human cardiac troponin T based on self-assembled monolayer. J Pharm Biomed Anal 2007,43(5):1744–1750.
Stevens RC, Soelberg SD, Near S, Furlong CE: Detection of cortisol in saliva with a flow-filtered, portable surface plasmon resonance biosensor system. Anal Chem 2008,80(17):6747–6751.
Cho HS, Park NY: Serodiagnostic comparison between two methods, ELISA and surface plasmon resonance for the detection of antibodies of classical swine fever. J Vet Med Sci 2006,68(12):1327–1329.
Vaisocherova H, Faca VM, Taylor AD, Hanash S, Jiang S: Comparative study of SPR and ELISA methods based on analysis of CD166/ALCAM levels in cancer and control human sera. Biosens Bioelectron 2009,24(7):2143–2148.
Chou SF, Hsu WL, Hwang JM, Chen CY: Development of an immunosensor for human ferritin, a nonspecific tumor marker, based on surface plasmon resonance. Biosens Bioelectron 2004,19(9):999–1005.
Mercey E, Sadir R, Maillart E, Roget A, Baleux F, Lortat-Jacob H, Livache T: Polypyrrole oligosaccharide array and surface plasmon resonance imaging for the measurement of glycosaminoglycan binding interactions. Anal Chem 2008,80(9):3476–3482.
Chung KF: Cytokines in chronic obstructive pulmonary disease. Eur Respir J Suppl 2001, 34:50s-59s.
Wallace AM, Sandford AJ, English JC, Burkett KM, Li H, Finley RJ, Muller NL, Coxson HO, Pare PD, Abboud RT: Matrix metalloproteinase expression by human alveolar macrophages in relation to emphysema. Copd 2008,5(1):13–23.
Feghali-Bostwick CA, Gadgil AS, Otterbein LE, Pilewski JM, Stoner MW, Csizmadia E, Zhang Y, Sciurba FC, Duncan SR: Autoantibodies in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2008,177(2):156–163.
Hartmann D, Felix K, Ehmann M, Schnolzer M, Fiedler S, Bogumil R, Buchler M, Friess H: Protein expression profiling reveals distinctive changes in serum proteins associated with chronic pancreatitis. Pancreas 2007,35(4):334–342.
Hsu HS, Chen TP, Wen CK, Hung CH, Chen CY, Chen JT, Wang YC: Multiple genetic and epigenetic biomarkers for lung cancer detection in cytologically negative sputum and a nested case-control study for risk assessment. J Pathol 2007,213(4):412–419.
Gygi SP, Rochon Y, Franza BR, Aebersold R: Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 1999,19(3):1720–1730.
Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG: Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 2002,1(4):304–313.
Barnes PJ, Chowdhury B, Kharitonov SA, Magnussen H, Page CP, Postma D, Saetta M: Pulmonary biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006,174(1):6–14.
Gora-Tybor J, Jamroziak K, Szmigielska-Kaplon A, Krawczynska A, Lech-Maranda E, Wierzbowska A, Jesionek-Kupnicka D, Blonski JZ, Robak T: Evaluation of circulating endothelial cells as noninvasive marker of angiogenesis in patients with chronic lymphocytic leukemia. Leuk Lymphoma 2009,50(1):62–67.
Sharma TS, Hughes J, Murillo A, Riley J, Soares A, Little F, Mitchell CD, Hanekom WA: CD8+ T-cell interleukin-7 receptor alpha expression as a potential indicator of disease status in HIV-infected children. PLoS ONE 2008,3(12):e3986.
Ueda K, Nakanishi T, Shimizu A, Takubo T, Matsuura N: Identification of L-plastin autoantibody in plasma of patients with non-Hodgkin's lymphoma using a proteomics-based analysis. Ann Clin Biochem 2008,45(Pt 1):65–69.
Holland M, Yagi H, Takahashi N, Kato K, Savage CO, Goodall DM, Jefferis R: Differential glycosylation of polyclonal IgG, IgG-Fc and IgG-Fab isolated from the sera of patients with ANCA-associated systemic vasculitis. Biochim Biophys Acta 2006,1760(4):669–677.
Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A: Serum microRNAs are promising novel biomarkers. PLoS ONE 2008,3(9):e3148.
Chen SY, Wang Y, Telen MJ, Chi JT: The genomic analysis of erythrocyte microRNA expression in sickle cell diseases. PLoS ONE 2008,3(6):e2360.
Lee KH, Su YD, Chen SJ, Tseng FG, Lee GB: Microfluidic systems integrated with two-dimensional surface plasmon resonance phase imaging systems for microarray immunoassay. Biosens Bioelectron 2007,23(4):466–472.
Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X: Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J Proteome Res 2006,5(6):1379–1387.
Fountoulakis M, Juranville JF, Jiang L, Avila D, Roder D, Jakob P, Berndt P, Evers S, Langen H: Depletion of the high-abundance plasma proteins. Amino Acids 2004,27(3–4):249–259.
Merrell K, Southwick K, Graves SW, Esplin MS, Lewis NE, Thulin CD: Analysis of low-abundance, low-molecular-weight serum proteins using mass spectrometry. J Biomol Tech 2004,15(4):238–248.
McIntosh M, Anderson G, Drescher C, Hanash S, Urban N, Brown P, Gambhir SS, Coukos G, Laird PW, Nelson B, Palmer C: Ovarian cancer early detection claims are biased. Clin Cancer Res 2008,14(22):7577–7579.
Thorpe JD, Duan X, Forrest R, Lowe K, Brown L, Segal E, Nelson B, Anderson GL, McIntosh M, Urban N: Effects of blood collection conditions on ovarian cancer serum markers. PLoS ONE 2007,2(12):e1281.
Bjorhall K, Miliotis T, Davidsson P: Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics 2005,5(1):307–317.
Vestergaard M, Tamiya E: A rapid sample pretreatment protocol: improved sensitivity in the detection of a low-abundant serum biomarker for prostate cancer. Anal Sci 2007,23(12):1443–1446.
Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, Kuick R, Misek DE, Cooke KR, Kitko CL, Weyand A, Bickley D, Jones D, Whitfield J, Reddy P, Levine JE, Hanash SM, Ferrara JL: A biomarker panel for acute graft-versus-host disease. Blood 2009,113(2):273–278.
Ludwig N, Keller A, Comtesse N, Rheinheimer S, Pallasch C, Fischer U, Fassbender K, Steudel WI, Lenhof HP, Meese E: Pattern of serum autoantibodies allows accurate distinction between a tumor and pathologies of the same organ. Clin Cancer Res 2008,14(15):4767–4774.
Moss AC, Jacobson GM, Walker LE, Blake NW, Marshall E, Coulson JM: SCG3 transcript in peripheral blood is a prognostic biomarker for REST-deficient small cell lung cancer. Clin Cancer Res 2009,15(1):274–283.
Leidinger P, Keller A, Ludwig N, Rheinheimer S, Hamacher J, Huwer H, Stehle I, Lenhof HP, Meese E: Toward an early diagnosis of lung cancer: an autoantibody signature for squamous cell lung carcinoma. Int J Cancer 2008,123(7):1631–1636.
Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, Lokshin AE, Lindell KO, Cisneros J, Macdonald SD, Pardo A, Sciurba F, Dauber J, Selman M, Gochuico BR, Kaminski N: MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med 2008,5(4):e93.
Karadag F, Karul AB, Cildag O, Yilmaz M, Ozcan H: Biomarkers of systemic inflammation in stable and exacerbation phases of COPD. Lung 2008,186(6):403–409.
Alagaratnam S, Mertens BJ, Dalebout JC, Deelder AM, van Ommen GJ, den Dunnen JT, t Hoen PA: Serum protein profiling in mice: identification of Factor XIIIa as a potential biomarker for muscular dystrophy. Proteomics 2008,8(8):1552–1563.
Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GL Jr: Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res 2002,62(13):3609–3614.
Cox B, Kislinger T, Emili A: Integrating gene and protein expression data: pattern analysis and profile mining. Methods 2005,35(3):303–314.
Fagan A, Culhane AC, Higgins DG: A multivariate analysis approach to the integration of proteomic and gene expression data. Proteomics 2007,7(13):2162–2171.
Yao J, Chang C, Salmi ML, Hung YS, Loraine A, Roux SJ: Genome-scale cluster analysis of replicated microarrays using shrinkage correlation coefficient. BMC Bioinformatics 2008, 9:288.
Ferguson GT, Enright PL, Buist AS, Higgins MW: Office spirometry for lung health assessment in adults: A consensus statement from the National Lung Health Education Program. Chest 2000,117(4):1146–1161.
Anthonisen NR, Connett JE, Kiley JP, Altose MD, Bailey WC, Buist AS, Conway WA Jr, Enright PL, Kanner RE, O'Hara P, et al.: Effects of smoking intervention and the use of an inhaled anticholinergic bronchodilator on the rate of decline of FEV1. The Lung Health Study. Jama 1994,272(19):1497–1505.
Franciosi LG, Page CP, Celli BR, Cazzola M, Walker MJ, Danhof M, Rabe KF, Della Pasqua OE: Markers of disease severity in chronic obstructive pulmonary disease. Pulm Pharmacol Ther 2006,19(3):189–199.
Acknowledgements
Prof P. O'Shea dedicates this paper to Dr. Rajeev Kalia, we hope our studies will eventually impact on his work on the respiratory frontline. JLR, EAML and RM are funded by the RCUK Basic Technology Programme. RAU is funded by The Jones' 1986 Charitable Trust.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JLR and RAU mainly wrote the manuscript, as well as the revision, and contributed equally to the study. LF conceived of the review and wrote part of the manuscript. POS conceived of the review and edited and wrote part of the manuscript. EAML, JC and RM helped to draft the manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Richens, J.L., Urbanowicz, R.A., Lunt, E.A. et al. Systems biology coupled with label-free high-throughput detection as a novel approach for diagnosis of chronic obstructive pulmonary disease. Respir Res 10, 29 (2009). https://doi.org/10.1186/1465-9921-10-29
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1465-9921-10-29