Abstract
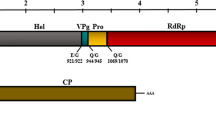
In this study, we describe the further genomic characterization of several pineapple mealybug wilt-associated virus (PMWaV) members of the genus Ampelovirus, family Closteroviridae, using high-throughput sequencing (HTS). PMWaV-2 is a necessary factor in the complex etiology of mealybug wilt disease of pineapple (MWP). The complete PMWaV-1 genome was described previously, but the genomes of PMWaV-2, PMWaV-3, and putative PMWaV-4 and PMWaV-5 species lack complete characterization. To further study MWP etiology through genomic characterization of PMWaVs, a cDNA library from total RNA of pineapple plants was generated and underwent HTS. A PMWaV-4 genome of 12,932 nucleotides was assembled from the HTS data. Proteins from the PMWaV-4 genome shared high amino acid similarity to those of PMWaV-1. The RNA-dependent RNA polymerase (RdRp), heat shock protein 70 homolog (HSP70h), and coat protein (CP) proteins of PMWaV-4 shared 88, 87, and 85% amino acid identity, respectively, to those of PMWaV-1 (AF414119) indicating PMWaV-4 is a distinct strain of PMWaV-1 but not a distinct ampelovirus species. Therefore, we suggest that PMWaV-4 be designated PMWaV-1 strain 4 (MN539275). Further, we used HTS to obtain a new isolate of PMWaV-1 (MN539276) and supplemented the available sequences of the genomes of PMWaV-2 (MN539272 and MN539273) and PMWaV-3 (MN539274) in their 5′-terminal regions.



Similar content being viewed by others
References
Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, Gruning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J (2016) The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research 44:W3–W10
Alvarez RA, Martin RR, Quito-Avila DF (2015) First report of pineapple mealybug wilt associated virus-1 in Ecuador. New Disease Reports 31:15
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Borroto EG, Cintra M, González J, Borroto C, Oramas P (1998) First report of a closterovirus-like particle associated with pineapple plants (Ananas comosus cv. Smooth Cayenne) affected with pineapple mealybug wilt in Cuba. Plant Disease 82:263–263
Carter W (1951) The feeding sequence of Pseudococcus brevipes-(ckl) in relation to mealybug wilt of pineapples in Hawaii. Phytopathology 41:769–780
Carter W (1952) Injuries to plants caused by insect toxins. II. The Botanical Review 18:680–721
Carter W (1963) Mealybug wilt of pineapple; a reappraisal. Annals of the New York Academy of Sciences 105:741–764
Dey KK, Borth WB, Melzer MJ, Wang ML, Hu JS (2015) Analysis of pineapple mealybug wilt associated virus-1 and -2 for potential RNA silencing suppressors and pathogenicity factors. Viruses 7:969–995
Dey K, Green JC, Melzer MJ, Borth W, Hu J (2018a) Mealybug wilt of pineapple and associated viruses. Horticulturae 4:52
Dey K, Sugikawa J, Kerr C, Melzer MJ (2018b) Air potato (Dioscorea bulbifera) plants displaying virus-like symptoms are co-infected with a novel potyvirus and a novel ampelovirus. Virus Genes 55:117–121
Gambley C, Steele V, Geering A, Thomas J (2008) The genetic diversity of ampeloviruses in Australian pineapples and their association with mealybug wilt disease. Australasian Plant Pathology 37:95–105
Gonsalves A, Sether D, Ullman D, Hu J (1992) Detection of pineapple closterovirus, a possible cause of mealybug wilt of pineapple. I International Pineapple Symposium 334:411–416
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature Biotechnology 29:644–652
Gunasinghe U, German T (1986) Association of virus-particles with mealybug-wilt of pineapple. Phytopathology 10:1073
Gunasinghe U, German T (1987) Further chacterization of virus associated with mealybug-wilt of pineapple. Phytopathology 12:1776
Gunasinghe U, German T (1989) Purification and partial characterization of a virus from pineapple. Phytopathology 79:1337–1341
Henschel R, Lieber M, Wu L, Nista P, Haas BJ, LeDuc RD (2012) Trinity RNA-Seq assembler performance optimization. In: Proceedings of the conference on extreme science and engineering discovery environment, article 45
Hernandez-Rodriguez L, Ramos-Gonzalez PL, Garcia-Garcia G, Zamora V, Peralta-Martin AM, Peña I, Perez JM, Ferriol X (2014) Geographic distribution of mealybug wilt disease of pineapple and genetic diversity of viruses infecting pineapple in Cuba. Crop Protection 65:43–50
Illingworth JF (1931) Preliminary report on evidence that mealy bugs are an important factor in pineapple wilt. Journal of Economic Entomology 24:877–889
Ito K (1962) Additional immunological evidence supporting the virus nature of mealybug wilt. Pineapple Research Institute News 10:158–162
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Larsen LD (1910) Diseases of pineapple. Bulletin of the Experiment Station of the Hawaii Sugar Planters’ Association. Division of Pathology and Physiology 10:1–72
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Molecular Biology and Evolution 25:1307–1320
Maliogka VI, Dovas CI, Katis NI (2008) Evolutionary relationships of virus species belonging to a distinct lineage within the Ampelovirus genus. Virus Research 135:125–135
Martelli GP, Agranovsky AA, Bar-Joseph M, Boscia D, Candresse T, Coutts RHA, Dolja VV, Hu JS, Jelkmann W, Karasev AV, Martin RR, Minafra A, Namba S, Vetten HJ (eds) (2011) Family Closteroviridae, ninth report of the international Commitee on taxonomy of viruses. Virus taxonomy. Elsevier-Academic Press, Amsterdam, p 996
Martelli GP, Abou Ghanem-Sabanadzovic N, Agranovsky AA, Al Rwahnih M, Dolja VV, Dovas CI, Fuchs M, Gugerli P, Hu JS, Jelkmann W, Katis NI, Maliogka VI, Melzer MJ, Menzel W, Minafra A, Rott ME, Rowhani A, Sabanadzovic S, Saldarelli P (2012) Taxonomic revision of the family Closteroviridae with special reference to the grapevine leafroll-associated members of the genus Ampelovirus and the putative species unassigned to the family. Journal of Plant Pathology 94:7–19
Melzer M, Karasev A, Sether D, Hu J (2001) Nucleotide sequence, genome organization and phylogenetic analysis of pineapple mealybug wilt-associated virus-2. Journal of General Virology 82:1–7
Melzer M, Sether D, Karasev A, Borth W, Hu J (2008) Complete nucleotide sequence and genome organization of pineapple mealybug wilt-associated virus-1. Archives of Virology 153:707–714
Peron FN, Calloni R, Ventura JA, Fernandes PMB (2019) Bioinformatics approach to the study of the molecular behaviour of mealybug wilt of pineapple. Acta Horticulture 1239:177–184
Sether D, Hu J (2002) Closterovirus infection and mealybug exposure are necessary for the development of mealybug wilt of pineapple disease. Phytopathology 92:928–935
Sether D, Ullman D, Hu J (1996) Detection of pineapple closterovirus in pineapple plants and mealybugs using monoclonal antibodies. Plant Pathology 45:829–836
Sether D, Liu X, Wang M, Zee F, Ullman D, Hu J (1997) Use of a tissue blotting immunoassay to examine the distribution of pineapple closterovirus in Hawaii. Plant Disease 81:1150–1154
Sether D, Ullman D, Hu J (1998) Transmission of pineapple mealybug wilt-associated virus by two species of mealybug (Dysmicoccus spp.). Phytopathology 88:1224–1230
Sether D, Karasev A, Okumura C, Arakawa C, Zee F, Kislan M, Busto J, Hu J (2001) Differentiation, distribution, and elimination of two different pineapple mealybug wilt-associated viruses found in pineapple. Plant Disease 85:856–864
Sether DM, Melzer MJ, Busto J, Zee F, Hu JS (2005a) Diversity and mealybug transmissibility of ampeloviruses in pineapple. Plant Disease 89:450–456
Sether D, Melzer M, Subere C, Hu J (2005b) Pineapple mealybug wilt associated viruses 1, 3, and 4, and grapevine leafroll associated viruses 4, 5, 6, and 9 are a distinct group in the genus Ampelovirus. In: XIII international congress of virology, Washington, DC. American Society for Microbiology, pp 121–122
Sether DM, Melzer MJ, Borth WB, Hu J (2009) Genome organization and phylogenetic relationship of pineapple mealybug wilt associated virus-3 with family Closteroviridae members. Virus Genes 38:414–420
Yu N, Luo Z, Fan H, Zhang Z, Li X, Wang J, Liu Z, He F (2015) Complete genomic sequence of a pineapple mealybug wilt-associated virus-1 from Hainan Island, China. European Journal of Plant Pathology 141:611–615
Acknowledgements
The research was supported in part by grants from the United State Department of Agriculture National Institute of Food and Agriculture, Hatch HAW09025-H (1001478), and the United State Department of Agriculture -Agricultural Research Service (58-5320-4-012). We appreciate Adriana E. Larrea-Sarmiento for her technical assistance.
Author information
Authors and Affiliations
Contributions
JG, MAR and JH planed and designed the experimental work. JG and MAR executed the experimental work. JG, AOV, and MAR conducted data analyses. JG, AOV, MM, IH, WB, MW, TM, and JH wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Section Editor: Michael Goodin
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Agarose gel electrophoresis of RT-PCR products for PMWaVs. Pineapple accessions samples were collected from the United States Department of Agriculture, Agricultural Research Service (USDA-ARS), National Clonal Germplasm Repository (NCGR) at the Daniel K. Inouye U.S. Pacific Basin Agricultural Research Center (PBARC) in Hilo HI, USA. (a) PMWaV-1, expected product size 590 bp; (b) PMWaV-2, expected product size 610 bp; (c) PMWaV-3, expected product size 499 bp; and (d) PMWaV-4, expected product size, 435 bp. Lanes: L, molecular ladder 100 bp; 1, HANA 17; 2, HANA 21; 3, HANA 46; 4, HANA 49; 5, HANA 156; 6, HANA 158; 7, HANA 160; 8, HANA 187; NTC, non-template control. (PNG 1544 kb)
Fig. S2
Agarose gel electrophoresis of RT-PCR products for partial fragments of RNA-dependent RNA polymerase (RdRp), heat shock protein 70 homolog (HSP70h), coat protein (CP) and ORF1a genes from PMWaV-4. (a) RT-PCR products of RdRP, HSP70 (HSP) and CP using primers 1944/1945, 1946/1947 and 1948/1949, respectively. Three technical replicates per gene were included in this RT-PCR assay. (b) RT-PCR products of partial fragments of ORF1a targeting positions 258-731 (primers 1932/1933), 94-553 (primers 1934/1935) and 798-1426 (primers 1936/1937) from a 1,522-nucleotide contig from a HTS assembly from HANA 160 infected with PMWaV-4. Six technical replicates were included in this RT-PCR assay. (PNG 180 kb)
Rights and permissions
About this article
Cite this article
Green, J.C., Rwahnih, M.A., Olmedo-Velarde, A. et al. Further genomic characterization of pineapple mealybug wilt-associated viruses using high-throughput sequencing. Trop. plant pathol. 45, 64–72 (2020). https://doi.org/10.1007/s40858-019-00330-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-019-00330-y