Abstract
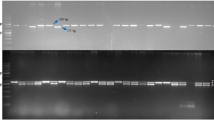
Pigeonpea is one of the most important pulse crops in the semi-arid tropical region, which is prone to several climatic uncertainties like unpredictable temperature, frequent drought and inconsistent rainfall. Additionally, during crop cycle pigeonpea also encounters a wide range of other biotic and abiotic constraints, ultimately leading to its fluctuating production and stagnant productivity. However, recently developed CGMS system has shown noteworthy impacts in enhancing pigeonpea productivity through exploitation of hybrid vigour. At present, A2-cytoplasm derived CGMS system has been well established in pigeonpea. Nevertheless, the commercial success of CGMS system relies largely on the continuous supply of genetically pure seeds of hybrids and corresponding parental lines. Traditionally, the genetic purity of seeds is guaranteed through conducting grow out test (GoT). In this context, DNA marker assays offer several advantages over conventional GoT especially in terms of time, space and money. Given its locus-specific and co-dominant nature, SSR or microsatellite marker is particularly suited for hybridity testing and purity assessment. Here we report a set of robust SSR markers, which could act as reliable molecular kit for ensuring the genetic purity of the CGMS-hybrid ‘IPH 09-5’ and its parental lines ‘PA 163A’ (A-or Male sterile-line) and ‘AK 261322’ (R- or Restorer-line).

Similar content being viewed by others
References
FAOSTAT (2011) http://www.faostat.fao.org
Odeny DA (2007) The potential of pigeonpea (Cajanus cajan (L.) Millsp.) in Africa. Nat Resour Forum 31:297–305
Varshney RK, Chen W, Li Y et al (2011) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89
Bohra A, Mallikarjuna N, Saxena K, Upadhyaya H, Vales MI, Varshney RK (2010) Harnessing the potential of crop wild relatives through genomics tools for pigeonpea improvement. J Plant Biol 37:1–16
Saxena KB, Sultana R, Mallikarjuna N, Saxena RK, Kumar RV, Sawargaonkar SL, Varshney RK (2010) Male-sterility systems in pigeonpea and their role in enhancing yield. Plant Breed 129:125–134
Pulses Newsletter IIPR, January–March 2013, pp8 (http://www.iipr.res.in/pdf/newsletter_2013_5june.pdf)
Yuan LP (1985) A concise course in hybrid rice. Hunan Science and Technology Publishing House, China, p 168
Saxena RK, Saxena KB, Varshney RK (2010) Application of SSR markers for molecular characterization of hybrid parents and purity assessment of ICPH 2438 hybrid of pigeonpea [Cajanus cajan (L.) Millspaugh]. Mol Breed 26:371–380
Yue B, Vick BA, Cai X, Hu J (2010) Genetic mapping for the Rf1 (fertility restoration) gene in sunflower (Helianthus annuus L.) by SSR and TRAP markers. Plant Breed 129:24–28
Bohra A, Dubey A, Saxena RK et al (2011) Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. BMC Plant Biol 11:56
Abdelnoor RV, Barros EG, Moreira MA (1995) Determination of genetic diversity within Brazilian soybean germplasm using random amplified polymorphic DNA techniques and comparative analysis with pedigree data. Braz J Genet 18:265–273
Sundaram RM, Naveenkumar B, Biradar SK et al (2008) Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica 163:215–224
Reddy BVS, Ashok Kumar A, Kaul SL (2008) Alternative cytoplasmic male sterility systems in sorghum and their utilization. In: Sorghum improvement in the new millennium. International Crops Research Institute for the Semi-Arid Tropics, Patancheru 502 324, Andhra Pradesh, India, pp. 132–144
Wan Z, Jing B, Tu J, Ma C, Shen J, Yi B, Wen J, Huang T, Wang X, Fu T (2008) Genetic characterization of a new cytoplasmic male sterility system (hau) in Brassica juncea and its transfer to B. napus. Theor Appl Genet 116:355–362
Rieseberg LH, Van Fossen C, Arias D, Carter RL (1994) Cytoplasmic male-sterility in sunflower-origin, inheritance, and frequency in natural populations. J Hered 85:233–238
Sivaramkrishnan S, Seetha K, Reddy LJ (2002) Diversity in selected wild and cultivated species of pigeonpea using RFLP of mtDNA. Euphytica 125:21–28
Ratnaparkhe MB, Gupta VS, Ven Murthy MR, Ranjekar PK (1995) Genetic fingerprinting of pigeonpea (Cajanus cajan (L.) Millsp) and its wild relatives using RAPD markers. Theor Appl Genet 91:893–898
Panguluri SK, Janaiah J, Govil JN, Kumar PA, Sharma PC (2005) AFLP fingerprinting in pigeonpea (Cajanus cajan L. Millsp.) and its wild relatives. Genet Resour Crop Evol 53:523–531
Yang S, Pang W, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A (2006) Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology (DArT). Theor Appl Genet 113:585–595
Bohra A, Saxena RK, Gnanesh BN, Saxena KB, Byregowda M, Rathore A, Kavi Kishor PB, Cook DR, Varshney RK (2012) An intra-specific consensus genetic map of pigeonpea [Cajanus cajan (L.) Millspaugh] derived from six mapping populations. Theor Appl Genet 125:1325–1338
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Odeny DA, Jayashree B, Ferguson M, Hoisington D, Crouch J, Gebhardt C (2007) Development, characterization and utilization of microsatellite markers in pigeonpea. Plant Breed 126:130–136
Saxena RK, Prathima C, Saxena K, Hoisington DA, Singh NK, Varshney RK (2010) Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.). Plant Breed 129:142–148
Varshney RK, Graner A, Sorrells E (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Burns MJ, Edwards KJ, Newbury HJ, Ford-Lloyd BV, Baggott CD (2001) Development of simple sequence repeat (SSR) markers for the assessment of gene flow and genetic diversity in pigeonpea (Cajanus cajan). Mol Ecol Notes 1:283–285
Odeny DA, Jayashree B, Gebhardt C, Crouch J (2009) New microsatellite markers for pigeonpea (Cajanus cajan (L.) Millsp.). BMC Res Notes 2:35
Singh NK, Gupta DK, Jayaswal PK et al (2011) The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol 21:98–112
Asif M, Rahman MU, Zafar Y (2006) Genotyping analysis of six maize (Zea mays L.) hybrid using DNA fingerprinting technology. Pak J Bot 38:1425–1430
Ali MA, Seyal MT, Awan SI, Niaz S, Ali S, Abbas A (2008) Hybrid authentication in upland cotton through RAPD analysis. Aust J Crop Sci 2:141–149
Acknowledgments
Financial support from the Indian Council of Agricultural Research (ICAR) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bohra, A., Singh, I.P., Yadav, A.K. et al. Utility of Informative SSR Markers in the Molecular Characterization of Cytoplasmic Genetic Male Sterility-Based Hybrid and its Parents in Pigeonpea. Natl. Acad. Sci. Lett. 38, 13–19 (2015). https://doi.org/10.1007/s40009-014-0288-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40009-014-0288-6