Abstract
Pigeonpea (Cajanus cajan) is an important grain legume of the Indian subcontinent, South-East Asia and East Africa. More than eighty five percent of the world pigeonpea is produced and consumed in India where it is a key crop for food and nutritional security of the people. Here we present the first draft of the genome sequence of a popular pigeonpea variety ‘Asha’. The genome was assembled using long sequence reads of 454 GS-FLX sequencing chemistry with mean read lengths of >550 bp and >10-fold genome coverage, resulting in 510,809,477 bp of high quality sequence. Total 47,004 protein coding genes and 12,511 transposable elements related genes were predicted. We identified 1,213 disease resistance/defense response genes and 152 abiotic stress tolerance genes in the pigeonpea genome that make it a hardy crop. In comparison to soybean, pigeonpea has relatively fewer number of genes for lipid biosynthesis and larger number of genes for cellulose synthesis. The sequence contigs were arranged in to 59,681 scaffolds, which were anchored to eleven chromosomes of pigeonpea with 347 genic-SNP markers of an intra-species reference genetic map. Eleven pigeonpea chromosomes showed low but significant synteny with the twenty chromosomes of soybean. The genome sequence was used to identify large number of hypervariable ‘Arhar’ simple sequence repeat (HASSR) markers, 437 of which were experimentally validated for PCR amplification and high rate of polymorphism among pigeonpea varieties. These markers will be useful for fingerprinting and diversity analysis of pigeonpea germplasm and molecular breeding applications. This is the first plant genome sequence completed entirely through a network of Indian institutions led by the Indian Council of Agricultural Research and provides a valuable resource for the pigeonpea variety improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pigeonpea or Red Gram (Cajanus cajan (L.) Millspaugh) is an important food legume for the tropical and subtropical regions of Indian subcontinent, South-East Asia and East Africa. It is a shrub with self-compatible cleistogamous flowers, but is often cross-pollinated by bees with 10–15% out crossing. The estimated size of pigeonpea genome packed in 11 chromosomes is 858 Mbp (Greilhuber and Obermayer 1998). It plays important role in food and nutritional security because it is a rich source of proteins, minerals and vitamins. Pigeonpea seeds are consumed mainly as split pea soups such as ‘Dal’ and ‘Sambar’ but a significant proportion is also consumed as green pea vegetable and whole grain preparations. Its leaves, seed husks and pod husks are used as animal feed (Fig. 1). Symbiotic bacteria (Bradyrhizobium) colonizing root nodules of pigeonpea fix atmospheric nitrogen up to 40 kg/ha in a cropping season and its deep root system improves soil structure and organic matter. Pigeonpea is unique among the legume crops as it is a woody shrub, therefore its stem and branches are used for firewood, fencing, thatch and making baskets by the rural population. Archeological evidence indicates that pigeonpea was domesticated in the eastern part of the Indian subcontinent along with rice and other important grain legumes, namely ‘Urd’ or black gram (Vigna mungo), ‘Mung’ or green gram (Vigna radiata) and ‘Kulthi’ or horse gram (Macrotyloma uniflorum) during prehistoric period (Fuller 2006). The world acreage of pigeonpea is 4.90 mha with annual production of about 4.22 mmt worth about 1.5 billion US dollars. India is the largest producer and consumer of pigeonpea (local names “Arhar”, “Tur”) with annual production of 3.07 mmt, followed by Myanmar (0.72 mmt) and Malawi (0.15 mmt) (FAOSTAT 2008).
Knowledge of the genetic basis of yield, quality and stress tolerance is important for genetic improvement of pigeonpea. Until a couple of years ago pigeonpea was considered an orphan legume crop but now substantial amount of genomic resources have been generated, largely owing to the efforts of Indo-US Agricultural Knowledge Initiative (AKI), NSF and GCP funded projects, (Varshney et al. 2009, 2010a; Dutta et al. 2011; Bohra et al. 2011). Pigeonpea cultivars have a narrow genetic base due to limited breeding efforts and poor utilization of wild pigeonpea species. Availability of genome sequence will accelerate the utilization of pigeonpea germplasm resources in breeding (Yang et al. 2006; Saxena 2008; Varshney et al. 2010b). Development of molecular markers tightly linked to the important agronomic traits is a prerequisite for undertaking molecular breeding in plants. But molecular basis of most agronomic traits in pigeonpea remains unexplored due to low level of DNA polymorphism in the primary gene pool and limited number of validated molecular markers (Ratnaparkhe et al. 1995; Yang et al. 2006; Odeny et al. 2009; Dutta et al. 2011; Bohra et al. 2011).
The aim of present study was: (a) to decode the pigeonpea genome by using next generation sequencing technologies and analyse its genes and repeat DNA contents; (b) generation of chromosome specific sequence by anchoring the sequence scaffolds to a high density reference molecular linkage map and its comparison with soybean genome; and (c) development of SSR markers for gene discovery and molecular breeding applications. Pigeonpea variety ‘Asha’ selected for this purpose is a popular variety with one of the highest breeder seed indents in India and is resistant to common diseases of pigeonpea, namely Fusarium wilt and sterility mosaic disease.
Materials and methods
Plant materials
Pigeonpea variety ‘Asha’ (ICPL87119) was used for genome sequencing and validation of newly designed HASSR markers. To identify informative HASSR markers, a set of 8 genotypes namely Asha, UPAS 120, HDM 04–1, Pusa Dwarf, H2004-1, Bahar, Maruti and TTB7 was screened for marker polymorphism. The seeds were obtained originally from IARI, New Delhi, ICRISAT Hyderabad, IIPR Kanpur and CCSHAU Hisar.
Genome sequence assembly and submission to NCBI GenBank
High quality genomic DNA was isolated from the leaves of a single plant of variety ‘Asha’ using CTAB method (Murray and Thompson 1980). Sequencing of 19 plates of whole genome shotgun libraries of short DNA fragments was carried out using GS-FLX Phase D chemistry, and 3 plates of paired end sequences from a library of 20 Kb long fragments of pigeonpea genomic DNA using GS-FLX Titanium chemistry (Margulies et al. 2005). Filtered high quality sequence reads were assembled using “Newbler GS De Novo assembler version 2.5.3” (Roeche Inc. Germany) with: Overlap minimum match length = 25 bp, Large genome = True, Number of CPU used = 0 (all), Exclude contigs of <500 bp. The GS Assembler is designed to compare all sequence reads in a pair wise fashion. Reads that overlap one another are joined into contigs. The consensus sequence for a contig is computed by taking an average of all aligned reads at a specific nucleotide position, the paired end reads were used for making scaffolds of sequence contigs. The large sequence contigs were quality checked and contaminating sequences were identified and removed. The quality check passed Fasta files containing 510,809,477 bp of pigeonpea genome sequence were further processed using command line software of NCBI to generate .sqn file (http://www.ncbi.nlm.nih.gov/HTGS/tbl2asninfo.html), which was submitted to GenBank as draft genome version 1 using Genomes Macro Send direct submission tool.
Gene annotation
The whole genome large sequence contigs were passed through FGENESH tool of MOLQUEST software (www.softberry.com) using Arabidopsis thaliana gene models as reference. From all predicted genes only those with size of >500 bp were taken for further analysis. The genes were BLAST searched against NCBI non-redundant database using optimized search parameters of gap opening penalty (G) = 4, gap extension penalty (E) = 1, mismatch score (q) = −1, match score (r) = 1, word size (W) = 11 and e-value <e−20 (Singh et al. 2004). Low complexity regions were included in the search. The BLAST search output was processed using BLAST Parser software (http://geneproject.altervista.org/). All the hits having bit scores of >100 and e values of <e−20 were tabulated in Microsoft Excel. Gene annotations were manually curated and categorized based on their functions. Details of pigeonpea transcriptome assemblies used for validation of predicted gene models is described earlier (Dutta et al. 2011). The predicted genes were manually curated with different keywords/phrases using auto filters to find R-like and defense response genes and categorize them into five main classes (Hulbert et al. 2001; The Rice Chromosomes 11 and 12 sequencing consortia 2005): (a) NBS-LRR (matching with NBS-LRR, but not with LZ-NBS-LRR and LRR, CC-NBS-LRR, Rp 1-d8, Lr10, Mla 1 and rust resistance), (b) LZ-NBS-LRR (matching with LZ-NBS-LRR, but not with NBS-LRR, CC-NBS-LRR, LRR and RPM1), (c) LRR-TM (matching with serine/threonine kinases and Cf2/Cf5 resistance), (d) miscellaneous category (matching with disease resistance, viral resistance, LRR, but not with NBS-LRR, CC-NBS-LRR, LZ-NBS-LRR), (e) defense response genes (matching with glucanases, chitinases and thaumatin like proteins). Similarly, genes for abiotic stress tolerance, lipid metabolism, sugar and starch biosynthesis, cellulose synthesis and transcription factors were also identified and categorized.
Annotation of transposable elements and repeats
Both De novo and homology based approaches were used for the identification of repeats in the large sequence contigs of pigeonpea genome. We used Repeat Modeler software pipeline for the construction of repeat library using RECON and Repeat Scout software (Benson 1999; Bao and Eddy 2002). Repeat Masker software was used for annotation using RMBLAST as search engine (Wootton and Federhen 1993; Lander et al. 2001; Waterston et al. 2002). Same strategy was used for the identification of repeats in genetically anchored scaffolds. We developed and added two different Perl scripts (Split masker, Masked table) in the Repeat Masker to break the large data set into individual files and simultaneously run the complete file in one go. Masked table script produced results on percentage of masked elements in each scaffold and exported it in Microsoft Ecxel.
For analysis of ribosomal RNA genes we downloaded all plant rDNA data from NCBI and used BLASTN search to find 28S, 18S and 5.8S rRNA genes in the pigeonpea genome. 5S rRNA genes were searched using a pigeonpea sequence obtained by cloning of Cot1 repeat fraction. tRNAscan software (Schattner et al. 2005; Lowe and Eddy 1997) was used for prediction of transfer RNA genes. The miRNA genes were identified using BLASTN search (e >1*10−5, top hits) of sequences present in the miRNA database, allowing no more than three mismatches (miRBAse release 17.0, Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Kozomara and Griffiths-Jones 2011). Rfam database (version 10.1, May 2011, Gardner et al. 2010; Griffiths-Jones et al. 2005) was used for identification of ribosomal, small nuclear and small nucleolar RNA genes. For snRNA only those families having 100% identity and e values of <0.001 were selected, whereas for snoRNA 80% identity and e values of <0.001 were selected.
Anchoring of sequence scaffolds to pigeonpea chromosomes
The sequence scaffolds were anchored to a high density linkage map of genic-SNP markers of an intra-species reference mapping population derived from Asha/UPAS120. The linkage map was based on two Illumina multiplex SNP assays of 1536-plex and 768-plex SNPs identified by comparing deep coverage transcriptome assemblies of the parental lines Asha and UPAS 120 (Dutta et al. 2011 and our unpublished results). The 59,681 sequence scaffolds assembled from the 454 GS-FLX sequence data were used to create a local database. Total 366 genic-SNP marker sequences genetically mapped to eleven pigeonpea chromosomes were BLASTN searched against this database at a cutoff bit score of ≥100 and e-value of <e−20. Gene density per 50 kb of anchored scaffolds was plotted for each chromosome at respective genetic map positions (cM) using Microsoft Excel. Anchored scaffolds were also scanned for the identification and annotation of RE using Repeat Modeler and Repeat Masker software, respectively. The percentage of RE in each scaffold was plotted against the gene density. The TE related genes in the scaffolds were identified using BLAST search in the NCBI-NR database.
Comparison between pigeonpea and soybean genomes
A total of 42,094 non-TE related genes were predicted from the pseudomolecules of twenty chromosomes of soybean (Glycine max) using the same approach as described above and a local database was created. Genes in the anchored scaffolds of pigeonpea were searched against this database using BLASTN with optimized search parameters (Singh et al. 2004). The output was parsed using BLAST Parser software (http://geneproject.altervista.org/) and tabulated in Microsoft Excel. Chromosomal positions of both pigeonpea and soybean genes were retained in the gene headers for analysis of synteny. Numbers of hits with bit scores ≥100 for each of the eleven pigeonpea chromosomes was counted in soybean and tabulated using Microsoft Excel. Similar comparison was made using single copy pigeonpea genes against the soybean chromosomes and a circular synteny map was plotted according to Krzywinski et al. (2009). To identify single copy genes a local database of all the predicted pigeonpea genes was created using ‘formatdb’ script of the NCBI local BLAST (Altschul, et al. 1990). All genes in the database were searched against themselves to find their copy numbers in the genome.
In silico mining, primer design and validation of genomic-SSR markers
All assembled contigs were screened for the presence of SSRs using MISA software (http://pgrc.ipk-gatersleban.de/misa). MISA created two types of files namely, 454AllContigs.fna.misa and 454AllContigs.fna.misa.statistics. MISA files were transferred to Microsoft Excel where SSRs were classified into mono-, di-, tri-, tetra-, penta- and hexa-nucleotide and compound repeats. The minimum repeat number was set at 10 for mono-, 6 for di-, and 5 for tri-, tetra-, penta- and hexa-nucleotides. Compound SSRs were defined as those loci having ≥2 SSRs interrupted by ≤100 bp of non-repetitive sequence. Class I SSRs with repeat lengths of ≥20 bp and hypervariable SSRs with repeat lengths of ≥50 bp were extracted according to Temnykh et al. (2001) and Singh et al. (2010), respectively. Nomenclature of markers HASSR1-HASSR437 using prefix H for hypervariable A for “Arhar” (pigeonpea) followed by SSR identification number was on the same pattern as describes earlier for pigeonpea genic-ASSR markers (Dutta et al. 2011). Primer pairs flanking the repeats were designed using Primer3 software (http://frodo.wi.mit.edu/). The target amplicon size was set to 100–260 bp, annealing temperature to 60°C, primer length to 20 bp and GC content to 50%. The primers were BLAST searched against the whole genome sequence to identify those with unique binding sites. For marker validation genomic DNA of eight pigeonpea genotypes was adjusted to a final concentration of 25 ng/μl. Total 437 genomic HASSR loci were first tested for PCR amplification using genomic DNA from Asha using PTC225 Gradient Cycler (Bio-Rad). PCR was carried out in 15 μl reaction volume containing 1.5 μl of 10× reaction buffer, 0.20 μl of 10 mM dNTPs (133 μM), 1.5 μl each of forward and reverse primers (10 pmol), 2.5 μl (62.5 ng) of template genomic DNA and 0.15 μl (0.75 U) of Taq DNA polymerase (Vivantis Technologies). The PCR cycling profile was: initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min., 55°C for 1 min., 72°C for 1 min and a final extension at 72°C for 7 min. Re-screening of primers that did not amplify at these conditions was done by sequentially decreasing the annealing temperature by 1°C; and for the primers producing multiple bands by sequentially increasing the annealing temperature by 1°C. The optimized SSR markers were then used for genotyping of eight varieties to check the level of polymorphism. PCR products were separated by electrophoresis in 4% Metaphor agarose gels (Lonza, Rockland USA) containing 0.1 μg/ml ethidium bromide in 1× TBE buffer at 130 V for 4 h, visualized and photographed in gel documentation system Fluorchem™ 5,500 (Alfa Innotech Crop., USA).
Results and discussion
Pigeonpea genome assembly
The aim of present study was to generate the first draft of pigeonpea genome sequence by making use of long sequence reads of 454 GS-FLX pyrosequencing ‘Phase D’ chemistry with modal read lengths of >550 bases. A total of 25,489,474 sequence reads with sequence information of 10,101,433,318 bp was generated. The primary sequence assembly included 21,102,008 sequence reads (82.79%) with 9.48 Gb sequence data, >10× coverage of the pigeonpea genome in 332,766 sequence contigs with consensus sequence of ~548 Mb. Of this, 192,089 contigs were larger than 500 bases with consensus sequence of ~511 Mb, average contig size of 2,661 bp, N50 contig size of 4,522 bp and largest contig size of 45,193 bp. After quality check (QC) 384 contig sequences were identified as bacterial contaminations and hence discarded. High quality of sequence assembly was evident from 97.9% (~500 Mb) of the consensus bases having Phred Quality scores of >40, reflecting an error rate of less than 1 in 10,000 bp (Ewing and Green 1998). Finally, 191,705 QC-passed large contigs with total 510,809,477 bp sequence. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AFSP00000000. The version described in this paper is the first version, AFSP01000000. The contigs were arranged into 59,681 scaffolds with the help of paired end sequences of 20 kb fragment library, covering ~458 Mb of genome sequence with average scaffold size of 7,679 bp, N50 size of 13,989 bp and the largest scaffold size of 177,971 bp. Thus 83.5% of the contig sequences were arranged in the scaffolds, and 16.5% still remained as singletons. The large sequence contigs, representing about 60% of the estimated 858 Mb size of the pigeonpea genome (Greilhuber and Obermayer 1998), were used for the analysis of genes and repeat contents of the genome and mining of SSR loci. In addition, 40.7 Gb of SOLiD mate pair sequence reads and 80.4 Gb of Illumina shotgun sequence reads have been generated for improving the genome coverage and sequence quality (Fig. 2). The published BAC paired end sequence data set is also available for improving the scaffolds (Bohra et al. 2011). Initial analysis showed 764.27 Mb coverage of the pigeonpea genome. However, present report describes analysis of the first draft using the 454 GS-FLX sequences only.
Gene content of the pigeonpea genome
The 454 GS-FLX large sequence contigs containing ~511 Mb of high quality sequence were used for gene prediction using FGENESH software. Total 59,515 genes were predicted with average gene size of 1,170 bp, largest gene size of 11,523 bp and the smallest gene size of 501 bp (Table 1). The average exon and intron sizes were 268 bp and 288 bp, respectively, which are comparable to soybean, the species most closely related to pigeonpea, for which genome sequence is available (Schmutz et al. 2010). The predicted coding sequences of the genes were compared with a high coverage transcriptome sequence assembly database including Sanger ESTs and 454-FLX transcriptome sequence assembly (TSA) contigs (Dutta et al. 2011). Approximately 99.9% of the predicted genes showed significant matches within the pigeon pea transcriptome database. Of the 59,515 predicted genes, 42,059 showed significant matches in the NCBI-NR database with bit scores higher than 100. Total 15,558 genes showed poor hits with bit scores lower than 100 and 2,147 genes showed no hits; therefore these sequences are unique to pigeonpea. Predicted genes were classified into different functional categories (Supplementary Table S1). Total 12,511 genes (21.02%) were TE-related and 27,441 genes (46.12%) were of unknown function. The unknown category includes 2,147 genes with no matches in the NCBI–NR database, 15,558 genes showing poor BLAST hits with bit scores <100 and 9,746 genes showing significant matches in the NCBI database with hypothetical category of genes. We added these 9,746 genes to the unknown function category because they show significant matches with our pigeonpea transcriptome database and hence are real genes showing expression. Only sixteen genes belonged to hypothetical category as they did not show significant match with any transcript sequence. The remaining 19,547 genes, 41.58% of the 47,004 protein coding genes, were those with known functions. Of these 6,180 were related to physiological traits, 1,213 for disease resistance and defense response, 3,601 for cellular transportation, 216 for stress response, 139 for protein synthesis, 453 for growth and development, 751 for DNA synthesis and repair and 6,994 genes for miscellaneous functions (Fig. 3).
Pigeonpea genome has a large number of 1,213 disease resistance (R-like) and defense response (DR) genes, which is 2.58% of all protein coding genes (Supplementary Table S1). These were divided into five classes based on sequence homology with the well established category of R-like and DR genes (Fig. 4). Total 98 DR genes were identified which belonged to three classes, namely chitinases (31 genes), glucanases (56 genes) and thaumatin-like proteins (11 genes). Out of 1,115 R-like genes, 219 (19.6%) belonged to miscellaneous category including genes for viral resistance, verticillium wilt resistance, bacterial blight resistance and genes containing LRR motif but without NBS, CC or LZ motifs. Of the total R-like genes the largest number of 459 genes (41.1%) showed homology to LRR-TM type genes, the second largest number of 436 genes (39.1%) showed homology to NBS-LRR type genes. Only one gene belonged to LZ-NBS-LRR category. The large number of disease resistance and defense response genes makes pigeonpea a hardy crop with fewer diseases.
Pigeonpea genome has 152 homologs of genes that have been implicated in abiotic stress tolerance in other plant species (Table 2). These include, 56 genes for heat shock proteins, 32 genes for glutathione-S-transferase (GST), 28 genes for trehalose-6-phosphate synthase (TPS), 8 genes for glutamine synthase (GS), 7 genes for water channel protein aquaporins and several transcription factors involved in abiotic stress response e.g. DREB, NAC and MYB genes (Supplementary Table S2).
Schmutz et al. (2010) identified 1,127 putative acyl lipid metabolism genes in the oilseed crop soybean. A similar analysis of genes for lipid metabolism in pigeonpea genome identified only 269 such genes, while soybean showed 536 genes (Table 2, Supplementary Table S3). Apart from the seed storage lipids these genes are involved in the metabolism of membrane lipids and various kinds of lipo-protein, glyco-lipid and mineral-lipid interactions. In contrast, pigeonpea genome has a higher number of 43 cellulose synthase genes as compared to only 37 genes in the soybean genome, which may be important for its woody plant architecture (Supplementary Table S4). Pigeonpea genome has 108 genes for the synthesis of various kinds of sugars, sugar transporters and starches including granule bound starch synthase, soluble starch synthase, starch branching and debranching enzymes. These have important implications for the grain yield and biomass accumulation (Supplementary Table S5). We identified 1,470 genes for different transcription factors and regulatory proteins in the pigeonpea genome (Table 2, Supplementary Table S6). These transcription factors play pivotal roles in the developmental regulation of gene expression and response of plants to various biotic and abiotic stresses. Most predominant transcription factors in the pigeonpea genome were AP2 domain-containing proteins, NAC domain containing proteins, WRKY transcription factors, Zinc finger proteins and MYB transcription factors.
Repeat elements in the pigeonpea genome
Identification and classification of repeat elements (RE) in large eukaryotic genomes is a challenging task that requires both de novo and homology based approaches (Lerat 2010). De novo analysis of RE using Repeat Modeler software revealed that pigeonpea, like other higher eukaryotic genomes, contains large proportion of repetitive DNA (Table 3). Repeat Modeler generated 1,811 different families of repeats known as the reference library. There were total 1,127,729 REs in the pigeonpea genome covering total 326,671,068 bp of sequence. Most REs (92.8%) were of interspersed type, comprising of Class I (Retro transposons), Class II (DNA transposons) or unclassified transposable elements. Simple direct repeats and low complexity repeats represented only 2.57% and 4.63% of the total RE, respectively. Homology based annotation using Repeat Masker identified REs belonging to six major categories, namely (a) LINEs including L1, R1, RTE-BovB; (b) LTR-retrotransposons including LTR, Caulimovirus, Copia, Gypsy; (c) DNA transposons including En-spm, Harbinger, hat-AC, hat-Tag1, hat-Tip100, Rc/Hiltron, MuDr, TcMAR-Pogo, RC/Hiltron; (d) Unclassified interspersed repeats; (e) Simple tandem repeats; and (f) Low complexity repeats. Similar to the other sequenced plant genomes, Gypsy and Copia type LTR-retrotransposons were the most predominant REs, constituting 16.02% and 6.10%, respectively. Interestingly, largest proportion (66.20%) of RE in the pigeonpea genome were unclassified and hence are unique to pigeonpea (Table 3). The total size of RE in the pigeonpea was 326.67 Mb which was 63.95% of the 511 Mb available genome sequence. The proportion of RE in the pigeonpea genome is higher than 23.90% in grape (Velasco et al. 2007), 25.03% in cucumber (Huang et al. 2009), 28.10% in Brachypodium (The International Brachypodium Initiative 2010), 34.79% in rice (IRGSP 2005) and 53.17% in papaya (Ming et al. 2008); similar to 61.47% in soybean (Schmutz et al. 2010), 67% in apple (Velasco et al. 2010), 64.13% in potato (The Potato Genome Sequencing Consortium 2011) and 62% in sorghum (Paterson et al. 2009); but lower than 84.20% in maize (Schnable et al. 2009) (Table 4).
DNA transposons constituted 2.99% of the pigeonpea genome, which is higher than apple (1.31%) but much lower than rice (37.25%), soybean (26.83%) and Brachypodium (16.98%). However, these proportions might be revised upwards after all the pigeonpea REs are classified. We identified 6,572 copies of hat-AC like families which has the highest frequency among the DNA transposons, followed by En-spm (5,166 copies) and TcMAR-Pogo (153 copies). Helitrons constituted only ~0.03% of the total RE in pigeonpea while sorghum showed the highest percentage of 1.3% (Table 4). The unclassified RE sequences represented the highest copy number of 623,425, covering 216 Mb of the available genome sequence. The interspersed repeats constituted 303 Mb (92.78%) of all RE in the pigeonpea genome, which was similar to soybean (95.53%). In contrast to the interspersed transposable elements, simple repeats and low complexity repeats contributed only 2.57% and 4.63% of the pigeonpea genome, respectively. These values were higher than 0.75% and 1.77% for the soybean genome (Schmutz et al. 2010).
Non-coding RNA genes in the pigeonpea genome
Genomes of higher plants contain thousands of copies of genes for non-coding RNA including rRNA, tRNA, miRNA, snRNA and snoRNA which play important role in the cellular protein synthesis machinery and regulation of expression of protein coding genes. In the pigeonpea genome we identified 35 copies of 28S rRNA genes, 66 copies of 18S rRNA (largest match of 2,346 bp in contig number 7,811) and 77 copies of 5.8S rRNA (largest match of 2,166 bp in contig number 77,111). We identified 270 copies of 5S rRNA genes using pigeonpea specific rDNA probes. We expect more copies of rRNA genes in the finished genome. The tRNAscan-SE software identified 671 tRNA genes. Of this, twenty were pseudogenes and two have undetermined anticodon isotypes. Remaining 649 tRNA genes have 50 different anticodons, representing all the twenty amino acids (Supplementary Table S7AB). The maximum number of genes were for leucine tRNAs (49), followed by serine (47), arginine (45) and glycine (45). Thirty six of the pigeonpea tRNA genes contain introns.
MicroRNAs (miRNAs) are important regulators of several biological processes like plant growth and development. These are 20–24 nucleotides in length. The 17th release of miRBAse database contains 19,724 mature miRNA sequences including 3,423 genes of plant origin. We identified 100 miRNA genes belonging to 32 different families in the pigeonpea genome (Fig. 5, Supplementary Table S8). Out of the 100 miRNA genes, 52 belong to miR829.1 family of Arabidopsis thaliana, which targets expression of three different proteins: (a) 3-ketoacyl-CoA reductase (b) P-glycoprotein and (c) central motor kinesin 1. There were four miRNA genes targeting ATP-sulfurylase and sulphate transporters (miR395), three genes each targeting Apetala2-like transcription factors (miR172) and F-box (miR393a) putative elements arein. There were twenty seven miRNA gene families with one or two copies of miRNA genes. Pigeonpea genome contains 226 snRNA genes showing homology in the Rfam database version 10.1 (May 2011). U6 family of snRNA showed the highest copy number of 97 genes, followed by U2 (48 genes) and U1 (34 genes) (Supplementary Table S9A). Further search with the Rfam database identified 335 sequences belonging to 90 families of snoRNA genes. The snoR71 family has the highest number of 166 genes, followed by snoRA7 and snoRD14 families having 10 genes each, the remaining families had 1–6 genes per family (Supplementary Table S9B).
Anchoring of pigeonpea sequence scaffolds to genetic map
We developed a high density intra-species reference genetic map of pigeonpea based on 366 genic-SNP markers (unpublished data). The 59,681 sequence scaffolds of pigeonpea genome were compared with the sequences of mapped genic-SNP markers and 347 (99.3%) of these showed matches with an equal number of scaffolds, covering total sequence of ~7.42 Mb. The anchored scaffolds provide genome wide nucleation points for the finishing of the pigeonpea genome and creation of large pseudomolecules for its eleven chromosomes. The 347 scaffolds were assigned to the eleven linkage groups of pigeonpea (Table 5). We predicted 1,041 genes in the anchored scaffolds, 63 of these genes were identified as TE-related genes and 26 genes did not show any hit in the database.
Out of the 7,424,371 bp of anchored scaffolds 1.697 Mb (23%) were RE which was less than half of the 63.95% RE in the whole pigeonpea genome, indicating that the anchored scaffolds represented gene-rich regions of the genome. Chromosome 10 showed the highest RE content (26.24%) while chromosome 11 showed the lowest RE content (15.43%). Anchored scaffolds represented only ~1.6% of the total ~458 Mb of assembled scaffolds, but they do provide a random sample of the genome and large number of nucleation points for the finishing of the genome. The average number of genes per 50 kb of scaffold sequence in the entire genome was 7.01 (Table 5). The gene density in the scaffolds was expected to be inversely proportional to the repeats density, which was true for many of the anchored scaffolds. For example, in chromosome 2, 3 and 5 we could clearly find this pattern for most of the scaffolds (Fig. 6). There was no uniform pattern for all the chromosomes, e.g. there was higher density of repeats in the middle portion of chromosomes 1, 5, 7 and 9, in one half of the chromosomes 4 and 10 and both the telomeric ends of chromosome 3. There was no clear difference in the repeat density along the lengths of chromosomes 2, 6, 8 and 11 (Fig. 6). In contrast, all twenty chromosomes of soybean have higher repeat density in the centromeric regions and higher gene density near the telomeres (Schmutz et al. 2010). The gene density in the anchored scaffolds of pigeonpea chromosomes was in the range of 6.31 to 9.97 per 50 kb. Chromosome 11 showed the highest gene density of 9.97% and lowest RE content of 15%. However, this picture may change as the size of scaffolds grows and we capture a high proportion of genome in the anchored scaffolds. We plan to merge the Illumina and Solexa data to increase the sequence coverage and BAC end sequence for increasing the size of anchored scaffolds.
Density of genes and repeat elements (TE) in the 347 anchored scaffolds on the eleven chromosomes of pigeonpea. Blue bars on the left side of each chromosome represent RE percentage in the scaffold and orange bars on the right side represent gene density per 50 kb. Red segments at the end of orange bars represent number of TE-related genes in the scaffold. Discontinuous blue bars indicate RE density of >40% whereas discontinuous orange bars represent gene density in excess of >10 genes per 50 kb
Comparative analysis of pigeonpea and soybean genomes
Pigeonpea and soybean belong to the same clade Millettieae of the plant family Fabaceae (Wojciechowski 2003). Both are important crop plants but have quite different plant architecture and seed composition. Pigeonpea is a shrub grown as annual crop that has high seed protein and starch contents but minimal oil content. Soybean on the other hand is an annual herb with seeds rich in oil and protein but low in carbohydrates. Therefore, we were interested to see the difference in the genome organization and gene content of the two species. The 47,004 protein coding genes of pigeonpea were compared with 42,094 protein coding genes of soybean using BLAST search with default parameters. Total 31,937 (67.94%) of the pigeonpea genes showed matches with soybean genes at a cutoff bit score of 100, whereas 9,067 genes were unique to pigeonpea. Similarly, out of 42,094 genes predicted in soybean 40,392 showed significant matches with pigeonpea genes, whereas 1,702 genes were unique to soybean. This shows that pigeonpea has significantly higher number of unique genes that differentiate it from soybean.
Conservation of synteny between pigeonpea and soybean was analysed on the basis of 347 genetically anchored scaffolds of pigeonpea. There are total 1,041 genes in the anchored scaffolds of which 512 are single copy genes. Number of matches with all genes and single copy genes of pigeonpea in twenty chromosomes of soybean are shown in Supplementary Table S10. Genes on each of the pigeonpea chromosomes showed matches with multiple soybean chromosomes but some soybean chromosomes showed significantly higher number of matches, and therefore are likely be syntenic. Another aspect to this analysis was comparison of all genes versus single copy pigeonpea genes which are shown to have a greater conservation of synteny between rice and wheat (Singh et al. 2007). Our comparison of all genes versus single copy pigeonpea genes with soybean also showed an improved visualization of synteny with single copy genes (Supplementary Table S10AB). Therefore, we focused on comparison of homology of single copy pigeonpea genes with the protein coding genes in twenty chromosomes of soybean (Fig. 7, Supplementary Table S10B). Chromosome 1 of pigeonpea showed matches with multiple soybean chromosomes even on the basis of single copy genes, but highest number of matches were found with chromosomes 8 and 5. Similarly, chromosome 2 of pigeonpea showed highest number of matches with chromosomes 19 and 10 of soybean. Chromosome 3 showed highest number of matches with chromosomes 13 and 15 of soybean. Chromosome 4 showed highest number of matches with chromosomes 12 and 13 of soybean. Chromosome 5 showed highest number of matches with chromosomes 13, 12 and 17 of soybean. Chromosome 6 showed highest number of matches with chromosomes 9 and 3 of soybean. Chromosome 7 showed highest number of matches with chromosomes 10 and 20 of soybean. Chromosome 8 of pigeonpea did not show high synteny with any specific chromosomes of soybean but it showed highest number of match with chromosomes 13 and 14. Chromosome 9 showed high number of matches with chromosomes 2, 12, 3, 11 and 16 of soybean. Chromosome 10 showed highest number of matches with chromosomes 18, 17 and 2 of soybean. Chromosome 11 of pigeonpea did not show high synteny with any specific chromosomes of soybean but highest numbers of matches were with chromosomes 14 and 18. A clear conservation of synteny was observed only with chromosomes 1, 3, 4 and 9 of pigeonpea with chromosomes 2, 5, 7, 8, 12, 13, 15 and 17 of soybean (Fig. 7). Chromosomes 2, 5, 6, 7 and 10 did not show clear synteny with any soybean chromosomes. Chromosomes 8 and 11 of pigeonpea did not show more than 10 matches with any of the soybean chromosomes (Fig. 7, Supplementary Table S10B). Low level of synteny between pigeonpea and soybean suggests that they might have only one genome in common and both are ancient amphipods. Their genomes have highly evolved after speciation from a common ancestral species; hence there is limited conservation of synteny between the two. This is in contrast to high conservation of macro synteny between rice and wheat, which separated about 50 mya (Singh et al. 2007).
Circular map of syntenic relationship between 11 pairs of pigeonpea chromosomes with 20 pairs of soybean chromosomes based on 512 single copy genes in the genetically anchored scaffolds of pigeonpea genome. The outer circles depict soybean chromosome bars showing proportion of gene matches with different chromosomes of pigeonpea and vice versa
Development and validation of hyper variable HASSR markers for pigeonpea
Pigeonpea genome was analysed to identify 1,89,895 SSR loci comprising of 100,373 mono-nucleotide, 49,325 di-nucleotide, 18,505 tri-nucleotide, 2,217 tetra-nucleotide, 512 penta-nucleotides, 815 hexa-nucleotide and 18,148 compound repeats (Table 6). Overall there is one SSR locus for every 2.88 kb of the pigeonpea genome sequence. Mononucleotide repeats are the most abundant class of SSRs in most genomes and pigeonpea was no exception to this. However, these do not serve as useful markers and excluding this category there was one SSR every 6.12 kb of the genome sequence. The frequency of SSR loci decreased successively with increasing size of the repeat unit from mono- to penta-nucleotide repeats, but frequency of hexa-nucleotide repeats was higher than penta-nucleotide repeats and compound repeats were much more abundant, comparable in frequency to the di-nucleotide repeats (Table 6). Among the two types of mono-nucleotide repeats, A/T were much more abundant than G/C (Supplementary Table S11). Among the di-nucleotide repeats, AT/AT was the most frequent while GT/AC and CG/CG were the least frequent. In the tri-nucleotide category AAT/ATT repeats were the most abundant while ACG/CGT and TAC/GTA were scarce. In the tetra-nucleotide category AAAT/ATTT was the most common motif whereas AAGG/CCTT, ACGT/ACGT, ACTA/TAGT and AGGA/TCCT were least frequent. In the penta-and hexa nucleotide categories also AT-rich repeats were more prevalent than the GC-rich repeats.
Search for class I SSRs (n ≥20 bp, Temnykh et al. 2001) and hyper variable HASSRs (n ≥50 bp, Singh et al. 2010) revealed that class I SSRs are most prevalent in the di-nucleotide category, whereas HASSRs are most abundant in the compound SSR category (Table 6). Based on the SSR length criteria 46,501 loci were classified as class I SSR and 11,711 of these were HASSR. All the SSR loci belonging to tetra-, penta-, hexa- and compound category were of class I SSR, while more than half (10,891) of the compound SSRs was of HASSR type. In contrast, mononucleotide repeats never reached a size of more than 50 bp, however this could be partly due to limitation of the 454 sequencing technology in dealing with large homopolymers. Due to their higher polymorphism longer SSR loci are more useful for routine genetic diversity analysis, fingerprinting, QTL mapping and molecular breeding applications in the laboratories lacking sophisticated fragment analysis and SNP genotyping platforms, but having simple agarose gel electrophoresis facility (Singh et al. 2010).
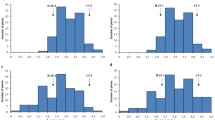
For wet lab validation we attempted to design PCR primers for 1,220 HASSR loci, taking 300 loci from the compound SSRs and all the loci from the remaining categories. But flanking primers could be designed successfully for amplification of only 530 of these loci, mainly due to location of the SSRs near one end of the sequence contigs. Surprisingly, no primer could be designed for the di-nucleotide category. Each of the designed primers was then compared with the whole genome sequence data to ensure that it bound to a unique position in the genome to prevent non-specific annealing. After this 93 loci were discarded due to multiple matches and primers were synthesized for 437 HASSR loci containing tri-, tetra-, penta- and hexa-nucleotide repeats as well as compound SSRs (Table 7). Details of validation results for the 437 HASSR markers, including primer sequences, Tm values, GC content and polymorphism level are shown in Supplementary Table S12. Total 358 primer pairs amplified a single PCR product of expected size and these were screened for polymorphism in a set of eight pigeonpea genotypes. We observed higher validation success rate of 81.92% for these genomic-SSR markers as compared to 72% success with genic-SSR markers described earlier (Dutta et al. 2011). HASSR markers showed 40.8% polymorphism (Table 7, Fig. 8), which is three times higher than 12.9% polymorphism observed with type I genic-SSR markers on the same set of eight genotypes (Dutta et al. 2011). Among the different categories of HASSR markers, complex SSRs showed the least polymorphism of only 6.5% (Table 7). This was discouraging because most of the HASSR loci belonged to this category (Table 6). The HASSR polymorphism was much higher than the earlier reported 28.40% polymorphism for BAC-end sequence derived genomic SSR markers obtained using high resolution capillary electrophoresis (Bohra et al. 2011). This underlines the high potential utility of the HASSR markers in pigeonpea molecular breeding.
The work presented here is the first draft of the whole genome sequence of pigeonpea and is the first report of a plant genome sequenced entirely in India. The 47,004 protein coding genes predicted in the pigeonpea genome are similar to that in soybean, potato and tomato, but significantly higher than Arabidopsis and rice. Ninety-nine point nine percent of the predicted genes were supported by RNA expression data, suggesting that these are true genes. A small proportion of genome scaffolds were genetically anchored with 347 mapped SNP markers which provide nucleation points for further finishing of the genome to large pseudomolecules of the eleven chromosomes. A comprehensive set of 46,501 Class I SSRs and 11,711 hypervariable HASSR loci were identified, and 437 HASSR markers were experimentally validated for amplification and higher rate of polymorphism. HASSR markers have high potential utility in the genetic diversity analysis, fingerprinting and molecular breeding for efficient utilization of pigeonpea germplasm resources in breeding improved varieties. The network partners under Indo-US AKI have already developed a EMS-mutagenized population and more than 24 recombinant inbred line populations for mapping of important agronomic traits including Fusarium wilt, sterility mosaic disease, flooding tolerance, seed size and number, plant type, drought tolerance and Dal (milling) quality of pigeonpea.
Abbreviations
- HASSR:
-
hypervariable ‘Arhar’ simple sequence repeats
- SNP:
-
single nucleotide polymorphism
- AKI:
-
Agricultural knowledge initiative
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bao Z, Eddy SR (2002) Automated de novo identification of repeats sequence families in sequenced genomes. Genome Res 12:1269–1276
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Bohra A, Dubey A, Saxena RK, Varma Penmetsa R, Poornima KN, Kumar N, Farmer AD, Srivani G, Upadhyaya HD, Gothalwal R, Ramesh S, Singh D, Saxena KB, Kavi Kishor PB, Singh NK, Town CD, May GD, Cook DR, Varshney RK (2011) Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea (Cajanus spp.). BMC Plant Biol 11:56
Dutta S, Kumawat G, Singh BP, Gupta DK, Singh S, Dogra V, Gaikwad K, Sharma TR, Raje RS, Bandhopadhya TK, Datta S, Singh MN, Fakrudin B, Kulwal P, Wanjari KB, Varshney RK, Cook DR, Singh NK (2011) Development of genic-SSR markers by deep transcriptome sequencing in pigeonpea [Cajanus cajan (L.) Millspaugh]. BMC Plant Biol 11:17
Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred II. Error probabilities. Genome Res 8:186–194
FAOSTAT 2008 [http://faostat.fao.org]
Fuller DQ (2006) Agricultural origins and frontiers in South Asia: a working synthesis. J World Prehist 20:1–86
Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, Bateman A (2010) Rfam: wikipedia, clans and the “decimal” release. Nucleic Acids Res 10:1093–1129
Greilhuber J, Obermayer R (1998) Genome size variation in Cajanus cajan Fabaceae: a reconsideration. Plant Syst Evol 212:135–141
Griffiths-Jones S (2004) The microRNA Registry. Nucleic Acids Res 32:D109–D111
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A (2005) Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 33:121–124
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, Ren Y, Zhu H, Li J, Lin K (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Lerat E (2010) Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity 104:520–533
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KLT, Salzberg SL, Feng L, Jones MR, Skelton RL, Murray JE, Chen C, Qian W et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya L.). Nature 452:991–996
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res 8:4321–4325
Odeny DA, Jayashree B, Gebhardt C, Crouch J (2009) New microsatellite markers for pigeonpea (Cajanus cajan (L.) millsp.). BMC Res Notes 2:35
Paterson AH, Bowers JE, Remy B, Inna D, Jane G, Gundlach H, Georg H, Uffe H, Therese M, Alexander P, Jeremy S, Manuel S, Haibao T, Xiyin W et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Ratnaparkhe MB, Gupta VS, Ven Murthy MR, Ranjekar PK (1995) Genetic finger printing of pigeonpea (Cajanus cajan (L.) Millsp.) and its wild relatives using RAPD markers. Theor Appl Genet 91:893–898
Saxena KB (2008) Genetic improvement of pigeonpea-a review. Trop Plant Biol 1:159–178
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–689
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, William N, Hyten DL, Qijian S, Thelen JJ, Jianlin C, Dong X et al (2010) Genome sequence of the palaeopolyploid Soybean. Nature 463:178–183
Schnable PS, Doreen W, Fulton RS, Stein JC, Fusheng W, Shiran P, Chengzhi L, Jianwei Z, Lucinda F, Graves TA, Patrick M, Reily AD, Laura C et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Singh NK, Raghuvanshi S, Srivastava SK, Gaur A, Pal AK, Dalal V, Singh A, Ghazi IA et al (2004) Sequence analysis of the long arm of rice chromosome 11 for rice–wheat synteny. Funct Integr Genomics 4:102–117
Singh NK, Vivek D, Kamlesh B, Singh Binay K, Chitra G, Archana S, Ghazi Irfan A, Mahavir Y, Awadhesh P, Rekha D, Singh PK, Harvinder S, Koundal Kirpa R, Kishor G, Trilochan M, Sharma Tilak R (2007) Single-copy genes define a conserved order between rice and wheat for understanding differences caused by duplication, deletion, and transposition of genes. Funct Integr Genomics 7:17–35
Singh H, Deshmukh RK, Singh A, Singh AK, Gaikwad K, Sharma TR, Mohapatra T, Singh NK (2010) Highly variable SSR markers suitable for rice genotyping using agarose gels. Mol Breed 25:359–364
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
The International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
The Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195
The Rice Chromosomes 11 and 12 sequencing consortia (2005) The sequence of rice chromosome 11 and 12, rich in disease resistance genes and recent gene duplication. BMC Biology 3: 1–18
Varshney RK, Close TJ, Singh NK, Hoisington DA, Cook DR (2009) Orphan legume crops enter the genomics era. Curr Opin Plant Biol 12:202–210
Varshney RK, Penmetsa RV, Dutta S, Kulwal PL, Saxena RK, Datta S, Sharma TR, Rosen B, Carrasquilla-Garcia N, Farmer AD, Dubey A, Saxena KB, Gao J, Fakrudin B, Singh MN, Singh BP, Wanjari KB, Yuan M, Srivastava RK, Kilian A, Upadhyaya HD, Mallikarjuna N, Town CD, Bruening GE, He G, May GD, McCombie R, Jackson SA, Singh NK, Cook DR (2010a) Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol Breed 26:393–408
Varshney RK, Thudi M, May GD, Jackson SA (2010b) Legume genomics and breeding. Plant Breed Rev 33:257–304
Velasco R, Andrey Z, Michela T, Cartwright DA, Alessandro C, Dmitry P, Massimo P, FitzGerald LM, Silvia V, Julia R, Giulia M, Diana I, Giuseppina C, Wardell B et al (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 12:31326
Velasco R, Zharkikh A, Jason A, Amit D, Alessandro C, Ananth K, Paolo F, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castellett S et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Waterston RH et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420(5):20–62
Wojciechowski MF (2003) Reconstructing the phylogeny of legumes (Leguminosae): an early 21st century perspective. In: Klitgaard BB, Bruneau A (eds) Advances in legume Systematics, Part 10, Higer level Systematics. Royal Botanic Gardens, Kew, pp 5–35
Wootton JC, Federhen S (1993) Statistics of local complexity in amino acid equences and sequence databases. Comput Chem 17:149–163
Yang S, Pang W, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A (2006) Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology (DArT). Theor Appl Genet 113:585–595
Acknowledgments
We are grateful to the Indian Council of Agricultural Research (ICAR) New Delhi for financial support through Indo-US Agricultural Knowledge Initiative (AKI) and Network Project on Transgenics in Crops (NPTC) projects. SD is grateful to the Council of Scientific and Industrial Research, Government of India for financial support (Grant no. 09/083/(0342)/2011/EMR-I)
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1
(XLS 8,103 kb)
Supplementary Table S2
(DOCX 45 kb)
Supplementary Table S3
(DOCX 48 kb)
Supplementary Table S4
(DOCX 23 kb)
Supplementary Table S5
(DOCX 37 kb)
Supplementary Table S6
(DOCX 183 kb)
Supplementary Table S7
(DOCX 131 kb)
Supplementary Table S8
(DOCX 39 kb)
Supplementary Table S9
(DOCX 19 kb)
Supplementary Table S10
(DOCX 23 kb)
Supplementary Table S11
(DOCX 17 kb)
Supplementary Table S12
(XLSX 85.7 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Singh, N.K., Gupta, D.K., Jayaswal, P.K. et al. The first draft of the pigeonpea genome sequence. J. Plant Biochem. Biotechnol. 21, 98–112 (2012). https://doi.org/10.1007/s13562-011-0088-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-011-0088-8