Abstract
Farinins are one of the oldest members of the gluten family in wheat and Aegilops species, and they influence dough properties. Here, we performed the first detailed molecular genetic study on farinin genes in Brachypodium distachyon L., the model species for Triticum aestivum. A total of 51 b-type farinin genes were cloned and characterized, including 27 functional and 24 non-functional pseudogenes from 14 different B. distachyon accessions. All genes were highly similar to those previously reported from wheat and Aegilops species. The identification of deduced amino acid sequences showed that b-type farinins across Triticeae genomes could be classified as b1-, b2-, b3-, and b4-type farinins; however, B. distachyon had only b3- and b4-type farinins. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) revealed that farinin genes are transcribed into mRNA in B. distachyon at much lower levels than in Triticeae, despite the presence of cis-acting elements in promoter regions. Phylogenetic analysis suggested that Brachypodium farinins may have closer relationships with common wheat and further confirmed four different types of b-type farinins in Triticeae and Brachypodium genomes, corresponding to b1, b2, b3 (group 1), and b4 (group 2). A putative evolutionary origin model of farinin genes in Brachypodium, Triticum, and the related species suggests that all b-type farinins diverged from their common ancestor ~3.2 million years ago (MYA). The b3 and b4 types could be considered older in the farinin family. The results explain the loss of b1- and b2-type farinin alleles in Brachypodium.







Similar content being viewed by others
References
Altenbach SB, Kothari KM (2004) Transcript profiles of genes expressed in endosperm tissue are altered by high temperature during wheat grain development. J Cereal Sci 40:115–126
Anderson OD, Greene FC (1997) The α-gliadin gene family. II. DNA and protein sequence variation, subfamily structure, and origins of pseudogenes. Theor Appl Genet 95:59–65
Anderson OD, Hsia CC, Adalsteins AE, Lew EJ-L, Kasarda DD (2001) Identification of several new classes of low-molecular-weight wheat gliadin-related proteins and genes. Theor Appl Genet 103:307–315
Badaeva ED, Friebe B, Zoshchuk SA, Zelenin AV, Gill BS (1998) Molecular cytogenetic analysis of tetraploid and hexaploid Aegilops crassa. Chromosome Res 6:629–637
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Benmoussa M, Vezina L-P, Pagé M, Yelle S, Laberge S (2000) Genetic polymorphism in low-molecular-weight glutenin genes from Triticum aestivum, variety Chinese Spring. Theor Appl Genet 100:789–793
Brenchley R, Spannagl M, Pfeifer M, Barker GLA, D’Amore R, Allen AM, McKenzie N, Kramer M, Bolser D, Kay S, Waite D, Gu Y, Huo N, Luo M-C, Sehgal S, Kianian S, Trick M, Bancroft I, Gill B, Anderson O, Dvorak J, Kersey P, McCombie R, Hall A, Mayer KFX, Edwards KJ, Bevan MW, Hall N (2012) Analysis of the allohexaploid bread wheat genome (Triticum aestivum) using comparative whole genome shotgun sequencing. Nature 491:705–710
Catalán P, Müller J, Hasterok R, Jenkins G, Mur LAJ, Langdon T, Betekhtin A, Siwinska D, Pimentel M, López-Alvarez D (2012) Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann Bot 109:385–405
Chen P, Wang C, Li K, Chang J, Wang Y, Yang G, Shewry PR, He G (2008) Cloning, expression and characterization of novel avenin-like genes in wheat and related species. J Cereal Sci 48:734–740
Chen P, Li R, Zhou R, He GY, Shewry PR (2010) Heterologous expression and dough mixing studies of a novel cysteine-rich Avenin-like protein. Cereal Res Commun 38:406–418
Chen GX, Lv DW, Li WD, Subburaj S, Yu ZT, Wang YJ, Li XH, Wang K, Ye XG, Ma W, Yan YM (2014) The α-gliadin genes from Brachypodium distachyon L. provide evidence for a significant gap in the current genome assembly. Funct Integr Genomics 14:149–160
Clarke BC, Phongkham T, Gianibelli MC, Beasley H, Bekes F (2003) The characterisation and mapping of a family of LMW-gliadin genes: effects on dough properties and bread volume. Theor Appl Genet 106:629–635
d’Aloisio E, Paolacci AR, Dhanapal AP, Tanzarella OA, Porceddu E, Ciaffi M (2010) The Protein Disulfide Isomerase gene family in bread wheat (T. aestivum L.). BMC Plant Biol 10:101
de Gregorio M, Armentia A, Díaz-Perales A, Palacín A, Dueñas-Laita A, Martín B, Salcedo G, Sánchez-Monge R (2009) Salt-soluble proteins from wheat-derived foodstuffs show lower allergenic potency than those from raw flour. J Agric Food Chem 57:3325–3330
Delcour JA, Joye IJ, Pareyt B, Wilderjans E, Brijs K, Lagrain B (2012) Wheat gluten functionality as a quality determinant in cereal-based food products. Annu Rev Food Sci Technol 3:469–492
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8
Drummond AJ, Bouckaert RR (2014) Bayesian evolutionary analysis with BEAST 2. Cambridge University Press, Cambridge
DuPont FM, Chan R, Lopez R, Vensel WH (2005) Sequential extraction and quantitative recovery of gliadins, glutenins, and other proteins from small samples of wheat flour. J Agric Food Chem 53:1575–1584
Dvořák J, Akhunov ED, Akhunov AR, Deal KR, Luo MC (2006) Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol Biol Evol 23:1386–1396
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Forde J, Malpica JM, Halford NG, Shewry PR, Anderson OD, Greene FC, Miflin B (1985) The nucleotide sequence of a HMW glutenin subunit gene located on chromosome 1A of wheat (Triticum aestivum L.). Nucleic Acids Res 13:6817–6832
Gao S, Gu YQ, Wu J, Coleman-Derr D, Huo N, Crossman C, Jia J, Zuo Q, Ren Z, Anderson OD, Kong X (2007) Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant Mol Biol 65:189–203
Gu YQ, Wanjugi H, Coleman-Derr D, Kong X, Anderson OD (2010) Conserved globulin gene across eight grass genomes identify fundamental units of the loci encoding seed storage proteins. Funct Integr Genomics 10:111–122
Guillon F, Larré C, Petipas F, Berger A, Moussawi J, Rogniaux H, Santoni A, Saulnier L, Jamme F, Miquel M, Lepiniec L, Dubreucq B (2012) A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J Exp Bot 63:739–755
Hammami R, Jouve N, Cuadrado A, Soler C, González JM (2011) Prolamin storage proteins and alloploidy in wild populations of the small grass Brachypodium distachyon (L.) P. Beauv. Plant Syst Evol 297:99–111
Hands P, Drea S (2012) A comparative view of grain development in Brachypodium distachyon. J Cereal Sci 56:2–8
Hasterok R, Marasek A, Donnison IS, Armstead I, Thomas A, King IP, Wolny E, Idziak D, Draper J, Jenkins G (2006) Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization. Genetics 173:349–362
Huo N, Vogel JP, Lazo GR, You FM, Ma Y, McMahon S, Dvorak J, Anderson OD, Luo MC, Gu YQ (2009) Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Mol Biol 70:47–61
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768
Jin M, Xie ZZ, Ge P, Li J, Jiang SS, Subburaj S, Li XH, Zeller FJ, Hsam SLK, Yan YM (2012) Identification and molecular characterisation of HMW glutenin subunit 1By16* in wild emmer. J Appl Genet 53:249–258
Kan YC, Wan YF, Beaudoin F, Leader DJ, Edwards K, Poole R, Wang DW, Mitchell RAC, Shewry PR (2006) Transcriptome analysis reveals differentially expressed storage protein transcripts in seeds of Aegilops and wheat. J Cereal Sci 44:75–85
Kasarda DD, Adalsteins E, Lew ELJ, Lazo GR, Altenbach SB (2013) Farinin: characterization of a novel wheat endosperm protein belonging to the prolamin superfamily. J Agric Food Chem 61:2407–2417
Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125:1198–1205
Larré C, Penninck S, Bouchet B, Lollier V, Tranquet O, Denery-Papini S, Guillon F, Rogniaux H (2010) Brachypodium distachyon grain: identification and subcellular localization of storage proteins. J Exp Bot 61:1771–1783
Laudencia-Chingcuanco DL, Vensel WH (2008) Globulins are the main seed storage proteins in Brachypodium distachyon. Theor Appl Genet 117:555–563
Li XH, Wang K, Wang SL, Gao LY, Xie XX, Hsam SLK, Zeller FJ, Yan YM (2010) Molecular characterization and comparative transcriptional analysis of LMW-m-type genes from wheat (Triticum aestivum L.) and Aegilops species. Theor Appl Genet 121:845–856
Li J, Wang SL, Cao M, Lv DW, Subburaj S, Li XH, Zeller FJ, Hsam SLK, Yan YM (2013) Cloning, expression, and evolutionary analysis of α-gliadin genes from Triticum and Aegilops genomes. J Appl Genet 54:157–167
Middleton CP, Senerchia N, Stein N, Akhunov ED, Keller B, Wicker T, Kilian B (2014) Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS One 9(3):e85761
Mur LA, Allainguillaume J, Catalán P, Hasterok R, Jenkins G, Lesniewska K, Thomas I, Vogel J (2011) Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytol 191:334–347
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Naghavi M, Ranjbar M, Zali A, Aghaei M, Mardi M, Pirseyedi S (2009) Genetic diversity of Aegilops crassa and its relationship with Aegilops tauschii and the D genome of wheat. Cereal Res Commun 37:159–167
Opanowicz M, Hands P, Betts D, Parker ML, Toole GA, Mills EN, Doonan JH, Drea S (2011) Endosperm development in Brachypodium distachyon. J Exp Bot 62:735–748
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer v1.6. Available from: http://beast.bio.ed.ac.uk/Tracer
Real A, Comino I, de Lorenzo L, Merchán F, Gil-Humanes J, Giménez MJ, López-Casado MÁ, Torres MI, Cebolla Á, Sousa C, Barro F, Pistón F (2012) Molecular and immunological characterization of gluten proteins isolated from oat cultivars that differ in toxicity for celiac disease. PLoS One 7:e48365
Salcedo G, Prada J, Aragoncillo C (1979) Low MW gliadin-like proteins from wheat endosperm. Phytochemistry 18:725–727
Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7:945–956
Shewry PR, Halford NG, Lafiandra D (2003) Genetics of wheat gluten proteins. Adv Genet 49:111–184
Song F, Cui CJ, Chen L, Sun YL, Wang FF, Hussain J, Li Y, Wang C, Wang C, Chen MJ, Wang YS, Yang GX, He GY (2012) Isolation and characterization of an endosperm-specific promoter from wheat (Triticum aestivum L.). Z Naturforsch C 67:611–619
Subburaj S, Chen G, Han C, Lv D, Li X, Zeller FJ, Hsam SLK, Yan YM (2014) Molecular characterisation and evolution of HMW glutenin subunit genes in Brachypodium distachyon L. J Appl Genet 55:27–42
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vogel J, Hill T (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27:471–478
Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D (2006a) EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor Appl Genet 113:186–195
Vogel JP, Garvin DF, Leong OM, Hayden DM (2006b) Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult 84:199–211
Vogel JP, Tuna M, Budak H, Huo N, Gu YQ, Steinwand MA (2009) Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol 9:88
Wang K, Han X, Dong K, Gao L, Li H, Ma W, Yan Y, Ye X (2010) Characterization of seed proteome in Brachypodium distachyon. J Cereal Sci 52:177–186
Wang S, Li X, Wang K, Wang X, Li S, Zhang Y, Guo G, Zeller FJ, Hsam SL, Yan Y (2011) Phylogenetic analysis of C, M, N, and U genomes and their relationships with Triticum and other related genomes as revealed by LMW-GS genes at Glu-3 loci. Genome 54:273–284
Wang S, Wang K, Chen G, Lv D, Han X, Yu Z, Li X, Ye X, Hsam SLK, Ma W, Appels R, Yan Y (2012) Molecular characterization of LMW-GS genes in Brachypodium distachyon L. reveals highly conserved Glu-3 loci in Triticum and related species. BMC Plant Biol 12:221
Wieser H, Kieffer R (2001) Correlations of the amount of gluten protein types to the technological properties of wheat flours determined on a micro-scale. J Cereal Sci 34:19–27
Xu JH, Messing J (2009) Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor Appl Genet 119:1397–1412
Yan Y, Zheng J, Xiao Y, Yu J, Hu Y, Cai M, Li Y, Hsam SL, Zeller FJ (2004) Identification and molecular characterization of a novel y-type Glu-D t 1 glutenin gene of Aegilops tauschii. Theor Appl Genet 108:1349–1358
Acknowledgments
The authors are grateful to Ms. Sarah Hancock for the language editing from the Department of Communications and Agricultural Education, Kansas State University Research and Extension. This research was financially supported by grants from the National Natural Science Foundation of China (31471485), Natural Science Foundation of Beijing City and the Key Developmental Project of Science and Technology, Beijing Municipal Commission of Education (KZ201410028031), International Science and Technology Cooperation Program of China (2013DFG30530), and the National Key Project for Transgenic Crops in China (2014ZX08009-003). This is contribution number 14-301-J from the Kansas Agricultural Experiment Station.
Conflict of interest
This manuscript has no financial or non-financial competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by: Andrzej Górny
Saminathan Subburaj and Nana Luo contributed equally to this work.
Electronic supplementary material
Below are the links to the electronic supplementary material.
Supplementary Figure 1
Chromosomal location of farinin-like genes from blast results in the Brachypodium genome (diploid line Bd21) from websites http://www.phytozome.net and http://www.brachypodium.org/. The black lines show the location of farinin-like genes in chromosomes with their corresponding megabase (Mb) position. The blue and orange triangles indicate the upward and downward directions of transcription of corresponding locus genes, respectively. (GIF 41 kb)
Supplementary Figure 2
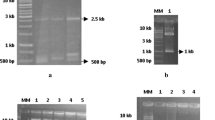
Characterization of b-type farinin genes isolated from Brachypodium. a Polymerase chain reaction (PCR) amplification of b3-type farinins. b PCR amplification of b4-type farinin genes. c 5′ promoter regions from b-type farinin genes. Lane 1, Chinese Spring; lane 2, Bd21; lane 3, Bd21-3; lane 4, Bd2; lane 5, Bd3; lane 6, Bd4; lane 7, Bd10; lane 8, Bd347; lane 9 Bd14; M is the 1-Kb DNA ladder. Target bands are indicated by arrows. (GIF 53 kb)
Supplementary Figure 3
Multiple alignments of b-type farinins from Triticum and Aegilops, along with cloned Brachypodium b-type farinins. (a, b, c). Various protein domains (signal, N-terminal, repeat: R1, R2, and C-terminal) are indicated by black, blue, and orange arrows. The significant residue variations help distinguish two groups (1 and 2) and four b (b1, b2, b3 and b4) farinins; those from Triticum, Aegilops, and Brachypodium are highlighted by yellow-shaded boxes in the alignment. (PDF 4284 kb)
Supplementary Figure 4
Comparison of the 5′ flanking region from five different Bd accessions (KF933340, KF933342, KF933336, KF933345, and KF933346), including diploid, hexaploid, and tetraploid genotypes with the T. aestivum promoter (JN622144) region. The detected cis-regulatory elements such as CAAT-box, TATA-box, endosperm motif, GCN4-like motif (GLM), RY motif, and G-box are indicated by boxes. (PDF 754 kb)
Supplementary Figure 5
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of b-type farinin genes. a–f Amplification efficiency and g–l dissociation melt curves for the amplicons of various reference genes (Ubiquitin, SAM-DC, and GAPDH), along with target genes (b3- and b4-type farinin) in CS (Chinese Spring) and Bd21 (B. distachyon) by qRT-PCR primers were tested. The non-template control (NTC) was confirmed by g–l gene-specific amplification and (M) single band amplification during qRT-PCR. (PDF 1185 kb)
Supplementary Figure 6
qRT-PCR analysis of b-type farinin genes. a–l Representation of Ct (threshold cycle) values for reference genes (Ubiquitin, SAM-DC, and GAPDH), along with target genes (b3- and b4-type farinins) during qRT-PCR analysis. (PDF 871 kb)
Supplementary Figure 7
Estimated divergence time between b-type farinin genes in Triticum family members based on aligned amino acid sequences using Bayesian Markov chain Monte Carlo (MCMC) analysis. A relaxed clock lognormal approach, Yule model of speciation process, and the WAG model of amino acid substitution were used. BEAST 2.1.3 was run for three independent times, each with 20,000,000 MCMC steps, and sampled once every 1000th generation. The molecular clock was calibrated using the divergence of B. distachyon–T. aestivum [35 ± 3 million years ago (MYA)] and A. speltoides–T. aestivum (0.87 ± 0.49 MYA). Estimated divergent time represented in MYA at each node. The green circle indicates the duplication event in the ancestors of both group 1 and group 2 farinins, and those within group 1 farinins are represented by a yellow circle. Farinin genes identified in this study are underlined. (PDF 93 kb)
Supplementary Table 1
Summary of blast results from the Phytozome and NCBI databases. The a- and b-type farinin genes cloned in this study were used to blast the Brachypodium genome database. Blast-annotated sequences are summarized here. (DOCX 16 kb)
Supplementary Table 2
List of primers used in this study to clone the farinin gene sequences and their expression under the heterologous Escherichia coli system. (DOCX 14 kb)
Supplementary Table 3
List of protein/peptide sequences identified from sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) bands of Fig. 4a (lanes 4 and 6) through LC-MS/MS analysis and searching the NCBInr database using MASCOT. (DOCX 14 kb)
Supplementary Table 4
Complete list of protein/peptide sequences identified from SDS-PAGE bands of Fig. 4a (lanes 4 and 6) through LC-MS/MS analysis and searching the NCBInr database using MASCOT. (XLS 51 kb)
Supplementary Table 5
List of primers designed and used for real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of mRNA expression studies of farinin genes. (DOCX 13 kb)
Supplementary Table 6
Absolute quantification of mRNA expression level of Ubiquitin, SAM-DC, and GAPDH genes in Brachypodium distachyon (Bd21) and Chinese Spring (CS) during the qRT-PCR analysis of target genes (b3- and b4-type farinins). Values are expressed as cDNA copies/μg of reverse transcribed total RNA. Data are shown as the mean ± standard deviation. (DOCX 17 kb)
Supplementary Table 7
The b-type farinin genes and its 5′ promoter from the current study and the previously reported HMW-GS, LMW-GS, and gliadins from Bd21 were used to blast against the unassembled Brachypodium genome sequences of Bd21-3, Bd3-1, Bd30-1, Bd1-1, BdKOZ, and BdTRC (ftp://brachypodium.org/brachypodium.org/NaturalVariation/). Blast-annotated sequences are summarized here. (XLSX 407 kb)
Rights and permissions
About this article
Cite this article
Subburaj, S., Luo, N., Lu, X. et al. Molecular characterization and evolutionary origins of farinin genes in Brachypodium distachyon L.. J Appl Genetics 57, 287–303 (2016). https://doi.org/10.1007/s13353-015-0316-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-015-0316-3