Abstract
The most commonly used normalization strategy for quantitative real-time reverse transcription-polymerase chain reaction (RT-qPCR) is to select a stable reference gene. However, to date, no suitable reference genes have been identified in sika deer antler tissues. Thus, the aim of this study was to identify the most stable gene or a set of genes to be used as reference genes for RT-qPCR analysis in sika deer antler tissues. We first selected candidate reference genes using sika deer antler gene expression data from an Illumina sequencing platform (Hiseq 2000); twenty-one reference genes from the antler tips of Chinese sika deer were selected to test for the normalization of expression levels during different growth stages. These genes were tested by RT-qPCR and ranked according to the stability of their expression using two different methods (implemented in geNorm and NormFinder). Based on different algorithms and analytical procedures, our results clearly indicate RPL40 and Gpx as the most stable reference genes of our pool.



Similar content being viewed by others
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR: a perspective. J Mol Endocrinol 34:597–601
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL et al (2009) The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
Calabrese R, Zampieri M, Mechelli R, Annibali V, Guastafierro T, Ciccarone F, Coarelli G, Umeton R, Salvetti M, Caiafa P (2012) Methylation-dependent PAD2 upregulation in multiple sclerosis peripheral blood. Mult Scler 18:299–304
Cankorur-Cetinkaya A, Dereli E, Eraslan S, Karabekmez E, Dikicioglu D, Kirdar B (2012) A novel strategy for selection and validation of reference genes in dynamic multidimensional experimental design in yeast. PLoS One 7:e38351
Coker JS, Davies E (2003) Selection of candidate housekeeping controls in tomato plants using EST data. Biotechniques 35:740–742, 744, 746 passim
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A (2007) Evidence based selection of housekeeping genes. PLoS One 2:e898
de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN (2005) Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 85:154–159
Foldager CB, Munir S, Ulrik-Vinther M, Søballe K, Bünger C, Lind M (2009) Validation of suitable housekeeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol 10:94
Goidin D, Mamessier A, Staquet MJ, Schmitt D, Berthier-Vergnes O (2001) Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal Biochem 295:17–21
Hamalainen HK, Tubman JC, Vikman S, Kyrölä T, Ylikoski E, Warrington JA, Lahesmaa R (2001) Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal Biochem 299:63–70
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Infante C, Matsuoka MP, Asensio E, Cañavate JP, Reith M, Manchado M (2008) Selection of housekeeping genes for gene expression studies in larvae from flatfish using real-time PCR. BMC Mol Biol 9:28
Kierdorf U, Kierdorf H (2011) Deer antlers: a model of mammalian appendage regeneration: an extensive review. Gerontology 57:53–65
Kužmová E, Bartoš L, Kotrba R, Bubenik GA (2011) Effect of different factors on proliferation of antler cells, cultured in vitro. PLoS One 6:e18053
Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J (2005) The TIGR gene indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33:D71–D74
Li C, Stanton JA, Robertson TM, Suttie JM, Sheard PW, Harris AJ, Clark DE (2007) Nerve growth factor mRNA expression in the regenerating antler tip of red deer (Cervus elaphus). PLoS One 2:e148
Macrae T, Sargeant T, Lemieux S, Hébert J, Deneault E, Sauvageau G (2013) RNA-Seq reveals spliceosome and proteasome genes as most consistent transcripts in human cancer cells. PLoS One 17:e72884
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Neville MJ, Collins JM, Gloyn AL, McCarthy MI, Karpe F (2011) Comprehensive human adipose tissue mRNA and MicroRNA endogenous control selection for quantitative real-time-pcr normalization. Obesity (Silver Spring) 19:888–892
Peltier HJ, Latham GJ (2008) Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 14:844–852
Pita-Thomas W, Fernández-Martos C, Yunta M, Maza RM, Navarro-Ruiz R, Lopez-Rodríguez MJ, Reigada D, Nieto-Sampedro M, Nieto-Diaz M (2010) Gene expression of axon growth promoting factors in the deer antler. PLoS One 5:e15706
Price J, Allen S (2004) Exploring the mechanisms regulating regeneration of deer antlers. Philos Trans R Soc Lond B Biol Sci 359:809–822
Price JS, Allen S, Faucheux C, Althnaian T, Mount JG (2005) Deer antlers: a zoological curiosity or the key to understanding organ regeneration in mammals? J Anat 207:603–618
Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR (2001) Beta-actin: an unsuitable internal control for RT-PCR. Mol Cell Probes 15:307–311
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Warner JR, McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34:3–11
Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M (2000) Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics 2:143–147
Zhou YH, Zhang YJ, Luo ZB, Fan YH, Tang GR, Liu LJ, Pei Y (2012) Selection of optimal reference genes for expression analysis in the entomopathogenic fungus Beauveria bassiana during development, under changing nutrient conditions, and after exposure to abiotic stresses. Appl Microbiol Biotechnol 93:679–685
Acknowledgments
This work was supported by the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (Grant No. 2011BAI03B02-2) and the Scientific and Technological Developing Scheme of Ji Lin Province (Grant No. 20140101124JC) and the National Nature Science Foundation of China (Grant No. 81072829 and 81303165).
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work; there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
RPKM (reads per kilobase of exon model per million mapped reads) value distribution of five factors related with bone development (PDF 54 kb)
Online Resource 2
RT-qPCR (quantitative real-time reverse transcription-polymerase chain reaction) assays used to evaluate the factors related with bone development (PDF 73 kb)
Online Resource 3
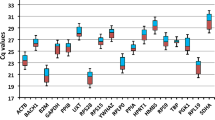
Variability of the candidate reference genes in the different samples (PDF 56 kb)
Online Resource 4
Expression profiles of five factors related with bone development. The most stable genes identified by geNorm (RPL40 and Gpx) and NormFinder (NADH) and the least stable gene GAPDH were used for normalization. For RPL40 and Gpx, the geometric mean was calculated and used for normalization. The relative expression levels are depicted as the standard deviation calculated from three biological replicates (PDF 120 kb)
Rights and permissions
About this article
Cite this article
Liu, M., Yao, B., Zhang, H. et al. Identification of novel reference genes using sika deer antler transcriptome expression data and their validation for quantitative gene expression analysis. Genes Genom 36, 573–582 (2014). https://doi.org/10.1007/s13258-014-0193-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0193-x