Abstract
Eukaryotic cells contain numerous iron-requiring proteins such as iron-sulfur (Fe-S) cluster proteins, hemoproteins and ribonucleotide reductases (RNRs). These proteins utilize iron as a cofactor and perform key roles in DNA replication, DNA repair, metabolic catalysis, iron regulation and cell cycle progression. Disruption of iron homeostasis always impairs the functions of these iron-requiring proteins and is genetically associated with diseases characterized by DNA repair defects in mammals. Organisms have evolved multi-layered mechanisms to regulate iron balance to ensure genome stability and cell development. This review briefly provides current perspectives on iron homeostasis in yeast and mammals, and mainly summarizes the most recent understandings on iron-requiring protein functions involved in DNA stability maintenance and cell cycle control.
Similar content being viewed by others
Introduction
In most eukaryotic cells, iron is necessary to facilitate the assembly of functional Fe-S cluster proteins, heme-binding proteins, and ribonucleotide reductases (RNRs) (Dlouhy and Outten, 2013; Heath et al., 2013). These iron-requiring proteins are abundantly present in mitochondria, cytosol, and nucleus; such proteins diversely function in electron transfer, ribosome maturation, DNA replication and repair, and cell cycle control (Kaplan et al., 2006; Ye and Rouault, 2010; White and Dillingham, 2012).
Unbalanced iron levels always affect the physiology of organisms. For instance, excess intracellular iron may result in the generation of reactive oxygen species (ROS), which can damage lipids, proteins, DNA; this adverse effect may eventually lead to genome instability and cell death in almost all organisms (Orrenius et al., 2011; Romero et al., 2014; Turrens, 2003). On the other hand, iron deficiency is extremely common in different species. Iron deficiency caused anemia is one of the major public health problems, particularly in children and pregnant women (Denic and Agarwal, 2007; Miller, 2013). In plants, the photosynthesis process is highly dependent on iron. Iron deficiency often reduces the amount of electron-transferring complexes, increases proteins involved in carbon fixation, and causes chlorosis (Lopez-Millan et al., 2013; Solti et al., 2008). In budding yeast Saccharomyces cerevisiae, iron deficiency leads to the dysfunction of iron-dependent enzymes, hemoproteins and Fe-S proteins, thereby altering glucose metabolism and biosynthesis of amino acid and lipid (Philpott et al., 2012; Shakoury-Elizeh et al., 2010).
Iron homeostasis in yeast and mammals
In eukaryotes, cellular iron homeostasis is achieved via strictly controlled systems for iron uptake at the plasma membrane and for eliciting balanced iron distributions among cellular compartments. In addition, the mammals should maintain systemic iron homeostasis by coordinately regulating iron absorption, storage and recycling, except keeping cellular iron balance.
Cellular iron homeostasis in yeast
Yeast cells acquire iron at the plasma membrane by utilizing high- and low-affinity iron uptake systems (Fig. 1). Under iron sufficient conditions, yeast cells mainly obtain iron through the low-affinity plasma membrane transporter Fet4. This process involves surface reductases (Fre1-Fre7), which can reduce Fe3+ to Fe2+ (Herbik et al., 2002; Holmes-Hampton et al., 2013; Wu et al., 2005; Yun et al., 2001). Under low iron conditions, yeast cells obtain iron via two independent high-affinity systems, particularly Fet3/Ftr1 complex and Arn1-4 proteins (Philpott, 2006; Rutherford et al., 2001; Yun et al., 2000a, b). The Fet3/Ftr1 proteins only deliver Fe2+ and their transcriptions are controlled by the Fe-responsive transcription factor Aft1 (Dlouhy and Outten, 2013; Hamza and Baetz, 2012). The Arn1-4 protein-dependent system becomes active in the absence of Fet3 protein, and this system is responsible for ferric siderophore uptake (Emerson et al., 2002; Heymann et al., 2000; Lesuisse et al., 1998; Yun et al., 2000a, b). When iron enters into the cytosol, it is present in a bioavailable form, namely, “labile iron pool” (LIP) (Fig. 1) (Muhlenhoff et al., 2010).
Iron uptake and utilization inside the cell. Yeast cells obtain iron through low-affinity (Fe-repleted condition, Fet4p) and high-affinity systems (low Fe condition, Fet3p/Ftr1p and Arn1-4 proteins). Both Fet4p and Fet3p/Ftr1p can only transport Fe2+, and these processes require the prior reduction of Fe3+ to Fe2+ by surface reductases (Fre1 to Fre7) (Herbik et al., 2002; Holmes-Hampton et al., 2013; Wu et al., 2005; Yun et al., 2001). The cytosolic “labile iron pool” is utilized by Fe-S proteins, hemoproteins, ribonucleotide reductases (RNRs), and other iron-requiring proteins that localize in different cellular compartments
Intracellular free iron is mainly stored in the vacuole, where it can be dynamically imported and exported by high- and low-affinity transporters (Fig. 2). The high-affinity vacuolar iron transport complex Fet5/Fth1 is a homologue of Fet3/Ftr1, and specifically responds to low iron (Amillet et al., 1996; Kaplan et al., 2006; Li and Kaplan, 1998; Li et al., 2008; Urbanowski and Piper, 1999). By contrast, the low-affinity vacuolar iron transporter Smf3 (DMT1 in mammals), which is a homologue of the cell membrane transporter Smf1, mainly functions in iron rich conditions (Portnoy et al., 2000; Portnoy et al., 2002). Interestingly, the Fet5/Fth1 complex and Smf3 are transcriptionally regulated by AFT1 and AFT2 (Li et al., 2008). Under Fe-deficient conditions, AFT1 and AFT2 activate the expressions of vacuolar iron exporter genes, particularly FET5/FTH1 and SMF3; as a result, cytosol iron is increased and vacuolar iron is decreased (Dlouhy and Outten, 2013). Moreover, iron storage in yeast requires vacuolar iron transporter Ccc1 and its expression is correlated with low iron. Activated AFT1 and AFT2 genes can induce CTH2, which subsequently binds to the 3′-untranslated region (UTR) of CCC1 and destabilizes its corresponding mRNA; the expression of CCC1 is then decreased (Li et al., 2001; Martinez-Pastor et al., 2013; Philpott et al., 2012). In addition, Yap5 has been indicated to function as an iron-responsive transcriptional activator that regulates vacuolar iron storage (Li et al., 2008).
Iron uptake, intracellular trafficking and regulation in S. cerevisiae. Iron uptake is performed at the plasma membrane by iron transporters. When iron enters into cytosol, it exists in Fe-S clusters in “labile iron pool” (Muhlenhoff et al., 2010), which is subsequently donated to cytosolic iron-dependent apoproteins through monothiol Grx3p/4p to form holoproteins. Meanwhile, the mitochondrial iron transporters Mrs3p/4p and the vacuolar iron transporter Smf3p, Ccc1p and Fet5p/Fth1p can also accept iron from Grx3p/4p. Grx3p/4p can interact with Fra2p and Aft1p/Aft2p, and the Grx3p/4p- bound Fe-S clusters may function as sensors for the cytosolic iron pool (Lill and Mühlenhoff, 2008; Muhlenhoff et al., 2010). In low-Fe condition, Aft1p can shuttle between the cytosol and the nucleus in an iron-responsive manner, and functions as a transcriptional activator of iron regulon genes, which subsequently activate high-affinity iron uptake systems (Berthelet et al., 2010; Lill et al., 2012)
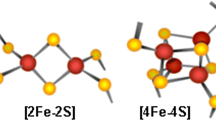
Aft1 localizes in the cytosol under iron rich conditions, whereas it accumulates in the nucleus when iron is low (Yamaguchi-Iwai et al., 2002; Ueta et al., 2012; Fig. 2). This nucleus-to-cytoplasm shuttling process is highly dependent on cytosolic monothiol glutaredoxins (Grx3 and Grx4), and the BolA-like protein Fra2 (Li et al., 2009). Grx3 (PICOT in mammals) and Grx4 are required to provide bioavailable iron for the assembly of many iron-containing holoproteins such as hemoproteins, Fe-S proteins, and RNR (Haunhorst et al., 2013; Mühlenhoff et al., 2010; Zhang et al., 2014a, b). Importantly, Grx3/4 can specifically interact with the CDC motifs in Aft1/2, and also can bind to Fra2 to form [2Fe-2S]-bridged heterodimers (Li et al., 2009; Ueta et al., 2007). The Fra2-Grx3/4 complex is possibly implicated in Aft1/2 sensing of the cellular iron status (Dlouhy and Outten, 2013). Under iron rich conditions, the Fra2-Grx3/4 complex binds to a Fe-S cluster, causing Aft1/2 oligomerization. Aft1 consequently shuttles to the cytosol through Msn5 and deactivates iron regulon. Under low iron conditions, the Fra2-Grx3/4 complex cannot inhibit Aft1/2 activities. Moreover, Aft1 moves to the nucleus and activates the iron regulon to increase cellular iron levels (Fig. 2) (Li et al., 2012; Lill et al., 2012; Poora et al., 2014).
Systemic and cellular iron homeostasis in mammals
Iron homeostasis in mammals includes systemic and cellular regulations, which have been recently summarized in detail (Andrews and Schmidt, 2007; Pantopoulos et al., 2012). Systemic iron regulatory processes occur in the following several steps: (1) iron absorption starts at duodenal enterocytes and functions in macrophage iron recycling and hepatocyte iron mobilization (Beaumont, 2010; Zhang, 2010); (2) intestinal cells then absorb iron via divalent metal transporter 1 (DMT1), which requires prior reduction of Fe3+ to Fe2+ by duodenal cytochrome b (DcytB) (Sharp and Srai, 2007; Pantopoulos et al., 2012); (3) spleenic reticuloendothelial macrophages control iron recycling from senescent red blood cells (Soe-Lin et al., 2009; Pantopoulos et al., 2012); (4) plasma transferrin (Tf) absorbs and circulates iron in the body (Pantopoulos et al., 2012); and (5) hepatic hormone hepcidin controls iron efflux by regulating the stability of ferroportin (Pantopoulos et al., 2012).
For cellular iron homeostasis, most mammalian cells acquire iron via Tf to form holo-Tf (Anderson and Vulpe, 2009; Dunn et al., 2007), which further binds to transferrin receptor-1 (TfR1) on the iron-consuming cell membrane. The holo-Tf-TfR1 complex is then internalized by receptor-mediated endocytosis (Lill et al., 2012) and acidified in the endosome. As a result, the release of Fe3+ from holo-Tf is facilitated (Zhao et al., 2010). The Fe3+ should be reduced to Fe2+ by a six-transmembrane epithelial antigen of the prostate 3 (Steap3) before this form of iron can be transported into the cytoplasm by DMT1 or transient receptor potential protein (TRPML1) (Zhang et al., 2012). Apo-Tf is then released from TfR1 and recycled back to the cell membrane to repeat another cycle (Pantopoulos et al., 2012). Thereafter, the newly acquired iron enters into the redox-active “labile iron pool” in the cytosol (Gkouvatsos et al., 2012; Pantopoulos et al., 2012). In addition, cellular iron balance is post-transcriptionally regulated by two iron regulatory proteins, namely, IRP1 and IRP2. Under Fe-deficient conditions, IRP1 and IRP2 specifically bind to iron-responsive elements in 3′- or 5′-UTR of the mRNA transcripts of TfR1, ferritin H chain (Fth1), ferritin L chain, or DMT1; as a result, these regulatory proteins are protected from degradation or their translation is inhibited (Anderson and Vulpe, 2009; Dunn et al., 2007; Kaplan and Kaplan, 2009; Muckenthaler et al., 2008).
Iron-requiring proteins and dna replication/repair
Numerous proteins involved in DNA replication and repair require iron as a cofactor. These proteins include the three DNA polymerases (Pol α, Pol δ and Pol ε), the DNA helicases (Rad3/XPD, Dna2, RTEL1, FANCJ and ChlR1), and DNA primase regulator subunit Pri2 (PRIM2 in mammals). Moreover, the eukaryotic RNR small subunit requires iron to form a diferric-tyrosyl radical cofactor (Fe III2- Y∙) to initiate nucleotide reduction.
DNA polymerases/primases and DNA replication
Although the eukaryotes contain diverse genomic sizes, the bulk of DNA synthesis is performed via three conserved polymerases: Pol α, Pol δ and Pol ε (Miyabe et al., 2011). Pol α tightly associates with DNA primases to initiate the synthesis of short RNA primers that are further utilized by Pol δ and Pol ε to synthesize the lagging and leading strands, respectively (Schumacher et al., 2000; Wang et al., 2004). Generally, eukaryotic DNA primases are heterodimeric enzymes with a small (PriS) and a large (PriL) subunit, in which the PriL subunit contains a conserved Fe-S domain necessary to initiate DNA replication (Kilkenny et al., 2013; Prakash and Prakash, 2002; Sauguet et al., 2010). Eukaryotes also possess Pol ζ, a B-family polymerase with lower fidelity than other polymerases; Pol ζ specifically functions in the extension step of translesion DNA synthesis (Acharya et al., 2006). All of these DNA polymerases and primases require a Fe-S cluster for the formation of active holoproteins, implying the importance of iron in maintaining genome integrity (Netz et al., 2012). Interestingly, the functions of these nuclear DNA polymerases depend on the cytosolic and the mitochondrial Fe-S protein biogenesis machineries, presumably because they act as sulfur donors for the Fe-S cluster in DNA polymerases (Rouault, 2012).
DNA helicases, DNA replication and repair
DNA helicase and helicase-nuclease enzymes, including XPD, Rad3, FancJ, ChlR1, RTEL1 and Dna2, preserve genome stability and are genetically associated with diseases characterized by DNA repair defects (Rudolf et al., 2006; Wu et al., 2009; Wu et al., 2012). Each of these proteins contains a conserved Fe-S cluster near the N-terminus, which is essential for helicase activities (Wu et al., 2012). XPD is classified as a SF2- DNA helicase and plays an important role in nucleotide excision repair (NER). FANCJ can catalytically unwind duplex DNA and G-quadruplex structures in an ATP hydrolysis-dependent manner (Wu et al., 2012). Clinically, patients with trichothiodystrophy (TTD) and Fanconi anemia completely lose helicase activities because of the relevant mutations in the Fe-S clusters of XPD and FancJ proteins, respectively (Coin et al., 2007; Fregoso et al., 2007). Moreover, site-directed mutagenesis of four conserved cysteines in Fe-S cluster of yeast XPD (Rad3) leads to defects in excision repair of UV photoproducts (Wu et al., 2009). The Fe-S cluster in the Rad3 helicase is necessary to induce the coupling of ATP hydrolysis with DNA translocation and to target helicase in the ss DNA-ds DNA junction (Pugh et al., 2008). ChlR1 is a DEAH/DEAD box-containing DNA helicase belonging to the FANCJ-like DNA helicase family. The mammalian and yeast ChlR1 proteins facilitate the establishment of sister chromatid cohesion and the maintenance of genomic stability. In Caenorhabditis elegans, CHL-1 is essential for normal development, fertility and chromosomal stability (Laha et al., 2011; Parish et al., 2006); RTEL specifically interacts with the shelterin complex and involves in telomere maintenance in mammals (Wu et al., 2012). These DNA helicases generally function together with other transcription factor II H (TFIIH) complex members such as XPG, XPB, p62, p52, p44, p34, p8/TTDA, Cdk7, cyclin H and MAT1 in human, and Rad3, Rad25, TFB1, SSL1, p55 and p38 in yeast (Sung et al., 1996).
The SF1 Dna2 helicase-nuclease, a protein implicated in double-strand break (DSB) end resection and Okazaki fragment processing, also contains a Fe-S cluster (Wu et al., 2012). Mutations in the Fe-S domain of Dna2 affect the ability of protein complexes to bind broken DNA, thereby impairing DNA replication; this result indicates the essential function of Fe-S in this process (Wu et al., 2012). Moreover, several other DNA helicases, such as DinG (E. coli), AddAB (E. coli), and DOG-1 (C. elegans), are implicated in DNA replication and repair (Wu et al., 2012).
Other iron-sulfur cluster proteins and DNA replication
The maturation of mitochondrial Fe-S proteins is carried out via the iron-sulfur cluster (ISC) assembly machinery, whereas cytoplasmic and nuclear Fe-S protein biogenesises depend on both the ISC and CIA (cytosolic iron-sulfur cluster assembly) machineries (Lill et al., 2012; Sipos et al., 2002). The components and biogenesis mechanisms of CIA pathway exhibit high degree of conservation in mammals and yeast. Yeast cells assemble a transiently bond [4Fe-4S] cluster on the Nbp35-Cfd1 scaffold (NUBP1-NUBP2 in mammals). This synthesis reaction further requires the electron transfer chain from Dre2 (CIAPIN1 in mammals) and its binding partner, the diflavin NADPH oxidoreductase Tah18 (NDOR1 in mammals). Thereafter, the transiently bound [4Fe-4S] cluster of Cfd1-Nbp35 is transferred to apoproteins, including apo-Cia1 (CIAO1 in mammals), apo-Nar1 (NARFL in mammals), apo-Cia2 and apo-MMS19, to form their corresponding holoproteins (Couturier et al., 2013; Zhang et al., 2008). The yeast MMS19 is necessary to transfer Fe-S clusters to target proteins and is also identified to affect DNA repair, chromosome segregation and heterochromatin silencing (Stehling et al., 2012). More importantly, both the human and yeast MMS19 proteins interact with numerous Fe-S proteins, including Pol δ, DNA primase, Dna2, XPD, RTEL1 and FANCJ (Gari et al., 2012). The stabilities of these Fe-S proteins are severely affected in the absence of MMS19 (Gari et al., 2012). Consistent with its essential role in DNA replication and repair, MMS19 deficiency in yeast or human cells exhibits increased sensitivity to hydroxyurea and S phase defect during cell cycle. Moreover, MMS19 can form a complex with two other CIA machinery proteins, namely, Cia1 and Cia2, suggesting that these CIA proteins are possibly involved in DNA replication and repair (Stehling et al., 2013).
The iron regulatory protein IRP1 possesses a [4Fe-4S] cluster. The inhibition of the IRP1 aconitase activity in L5178Y mouse lymphoma cells can increase “labile iron pool” levels. The increased iron burden in LIP leads to exacerbated hydrogen peroxide-induced genotoxicity in L5178Y cells (Lipinski et al., 2005). The stable IRP1 knockdown by shRNA interference in HL60 radiosensitive cells causes radioresistance to linear energy transfer gamma rays, but a more rapid DNA DSB repair. The mechanism of radioresistance is possibly related to the attenuated free radical-induced cell death (Haro et al., 2012).
RNR, iron, DNA replication and DNA repair
RNRs are enzymes that use radical chemistry to reduce ribonucleotides to synthesize deoxyribonucleotides (dNTPs), thereby generating the necessary precursors of DNA replication and repair (Zhang et al., 2014a, b). Imbalanced dNTP pools usually lead to increased DNA mutations, DNA breaks and cell death by enhancing misincorporation and by inhibiting the proofreading function of DNA polymerases (Kumar et al., 2010; Zhang et al., 2014a, b). Eukaryotic RNRs comprise the large subunits (α or R1) and small subunits (β or R2). Similar to Fe-S proteins, the RNR small subunits also require iron to sustain a diferric tyrosyl radical (Fe III2- Y∙) cofactor. Cells depleted of Grx3/4 exhibit deficiencies in RNR nucleotide reduction activity, but the mechanism remains to be elucidated (Zhang et al., 2008; Li et al., 2009; Netz et al., 2010, Ueta et al., 2012). Interestingly, the recent studies have shown that the depletion of Dre2 affects both RNR gene transcriptions and mRNA turnover by activating Aft1/Aft2-controlled iron regulon (Zhang et al., 2014a, b). The RNR subunit protein levels are tightly regulated by the DNA damage checkpoint. For instance, the mammalian small subunit RRM2 is degraded via an ubiquitin-mediated mechanism when cells complete DNA replication and/or repair. This process is mediated through two E3 ubiquitin ligase complexes, namely, the Skp1/Cullin/F-box (SCF) and the anaphase-promoting complex (APC) (Chabes et al., 2003b; D’Angiolella et al., 2012). In response to DNA damage, mammalian cells increase the transcription of p53R2 (RRM2B), which further forms an active RNR holoenzyme with RRM1 to facilitate DNA repair by activating the ATM/ATR-CHK checkpoint pathway (Harper and Elledge, 2007; Nakano et al., 2000). Similarly, the expressions of yeast RNR genes are induced via the activation of the Mec1-Rad53-Dun1 damage checkpoint kinase cascade, particularly RNR3 (Zhang et al., 2014a, b).
Hemoproteins and DNA stability
Heme commonly serves as the prosthetic group for hemoproteins, such as hemoglobin, myoglobin, cytochromes and nitric oxide synthase. These hemoproteins are involved in oxygen transport, oxidative catalysis and electron transport (Rae and Goff, 1998; Brown et al., 2004; Pamplona et al., 2007; Girvan and Munro, 2013). In addition, heme is important for systemic iron homeostasis in mammals, as it is present in many normal dietary sources (Pamplona et al., 2007). A number of heme transporters (FLVCR, ABCG2/BCRP, ABCB6, ABCB7 and ABCB10, PCFT/HCP1, HRG-1 and HRG-4) and heme-binding transcription factors (Bach1, NPAS2 and Rev-erb) are reportedly involved in heme metabolism and regulation (Severance and Hamza, 2009; Yang et al., 2008). Many hemes are enzymatically degraded by their degradation systems, such as heme oxygenases (HO, including HO-1, 2, and 3) and microsomal cytochrome P450 reductase. A considerable amount of hydrogen peroxide (H2O2) is produced during heme degradation, which may cause cellular toxicity and DNA damage (Quincozes-Santos et al., 2013; Wagener et al., 2003).
The disruption of hemoproteins, such as cytochromes b 5 and nitric oxide synthase, possibly increases ROS production. Cytochromes b 5 is a membrane bound hemoprotein and generally serves as an electron carrier in several oxidative reactions of reductases, such as NADH-cytochrome b5 reductase (Reid et al., 2013; Schenkman and Jansson, 2003; (Vergeres and Waskell, 1995), NADPH-cytochrome P450 reductase (Gan et al., 2009; Pyrih et al., 2014), and fatty acid desaturases involved in lipid and cholesterol biosynthesis (Reddy et al., 1977; Keyes and Cinti, 1980; Larade et al., 2008). The yeast Irc21, which shares similar heme-binding sites with Cyb5, reportedly functions in chromatin remodeling and the increase of DNA damage foci (Alvaro et al., 2007). This result indicates that Irc21 may be involved in DNA replication process, and the assumption is supported by the genetic interactions of Irc21 with several DNA damage- and repair-related proteins, such as Pri2, Pol12, Dia2, Rad17, MMS22 and CDC13 in Saccharomyces genome database (SGD). Moreover, cytochrome c is also a small hemoprotein involved in apoptosis and cell death (Jiang et al., 2004).
Anemia and dna stability
Anemia occurs as a result of numerous underlying causes and can be classified into different types based on the morphologies of red blood cells, discernible clinical spectra and etiologic mechanisms (Shah and Agarwal, 2013). As the most common form of anemia, iron deficiency anemia increases nuclear DNA damage in adults, as demonstrated by an increased DNA damage in anemic subjects (Aslan et al., 2006). Conversely, the results of iron nutritional deficiency in rats do not affect DNA stability or lipid peroxidation (Diaz-Castro et al., 2008). Studies have also indicated that dietary iron-deficient anemia induces various metabolic changes and even apoptosis in rat liver (Kamei et al., 2010). Moreover, fanconi anemia, a genetic disorder, is caused by defects in a cluster of proteins responsible for DNA repair. Studies have shown that eight of these proteins (FANCA, -B, -C, -E, -F, -G, -L and -M) assemble to form a core protein complex in the nucleus. Their assemblies are activated by replicative stresses, particularly DNA cross-linking agents (mitomycin C or cisplatin) and ROS (Deans and West, 2009). The deficiency of several ribosomal proteins (RP) can cause diamond blackfan anemia (DBA), which is a genetic syndrome characterized by red blood cell aplasia. RP-deficient zebrafish and human hematopoietic activate the ATR/ATM/CHK1/2/p53 pathway (Danilova et al., 2014).
Iron-requiring proteins and cell cycle control
Iron is a major regulator of cell cycle by inhibiting the formation or activities of the cyclin and cyclin-dependent kinase complexes. Intracellular iron disruption by chelators causes cell cycle arrest, particularly in G1 and S phases (Fu et al., 2007; Siriwardana and Seligman, 2013).
Cyclin, iron and cell cycle
The yeast cell cycle is mainly regulated by CDK (Cdc28), together with two other cyclin families: Cln1–3 and Clb1–6 (Nasmyth, 1993; Mendenhall and Hodge, 1998). Cdc28 associates with different cyclin proteins to govern cell cycle issues. For instance, Cln1/Cdc28 and Cln2/Cdc28 function in budding generation and spindle pole body duplication; Cln3/Cdc28 controls the size of newly formed cells (Chen et al., 2000). Clb1/Cdc28 and Clb2/Cdc28 are involved in mitosis; Clb3/Cdc28 and Clb4/Cdc28 are implicated in DNA replication and spindle formation, Clb5/Cdc28 and Clb6/Cdc28 are required for DNA replication (Mendenhall and Hodge, 1998; Ofir and Kornitzer, 2010; Vohradsky, 2012).
Constant AFT1 expression results in an increased iron uptake, thereby leading to cell cycle arrest at the start of G1 regulatory point (Philpott et al., 1998). The expression of the G1-specific cyclins (Cln1 and Cln2) is decreased when yeast cells are exposed to iron rich conditions, which may account for the arrest (Philpott et al., 1998; Yu et al., 2007).
In human, cyclins and CDK also control cell cycle progression. Intracellular iron depletion by some chelators causes allosteric inhibition of cyclin-A, cyclin-E, Cdc2 and Cdk2, resulting in cell cycle arrest in G1 and S phases (Renton and Jeitner, 1996; Yu et al., 2000). In some cases, intracellular iron depletion reduces cyclin-D and Cdk4 protein levels and alters retinoblastoma protein phosphorylation (Yu et al., 2000). This G1/S arrest further indicates the essential roles of iron in cell cycle progression, growth and division. Under some iron deficient conditions, a G2/M arrest has also been detected (Renton and Jeitner, 1996). Furthermore, several iron chelators evidently exhibit strong anticancer activities by inducing cell cycle arrest and apoptosis (Rao, 2013). However, involved mechanisms are barely known; as such, the relationships between iron chelators and structural activities should be understood.
Iron-sulfur cluster proteins and cell cycle
In yeast, the expressions of iron transporter genes FET3/FTR1 are tightly regulated by cell cycle and reach the peaks during M and M/G1 phases (Spellman et al., 1998). Two tah18 temperature-sensitive (ts) mutants, namely, tah18-5I5 and tah18-5H8, exhibit a prolonged S phase and a delay at the G1/S boundary, respectively (Zhang et al., 2014a, b). MMS19 gene silencing in human cells leads to genotoxic stress sensitivity (Stehling et al., 2012) and G1 phase arrest in cell cycle under limited nucleotide pool conditions, suggesting that DNA replication is impaired in these cells (Gari et al., 2012).
In human, the upregulation of CIAPIN1 results in significant inhibition of the CCRCC-derived cell growth in vitro and in vivo with G1 cell cycle arrest (He et al., 2009). CIAPIN1-induced growth suppression reduces protein levels of cyclin D1, cyclin E, Cdk2, Cdk4, p-Rb and VEGF (He et al., 2009). Moreover, Grx3 is critical for cell cycle progression during embryogenesis in mouse. Cells depleted of Grx3 undergoes normal DNA replication during the S phase but exhibit impaired growth and cell cycle progression at the G2/M phase (Cheng et al., 2011).
Iron-requiring proteins and cell cycle
In budding yeast, information on the hemoproteins involved in cell cycle is limited. However, heme and hemoproteins have been implicated in controlling the expressions of cell cycle regulators and cell growth in mammals. Heme synthesis inhibition causes cell cycle arrest in S phase, upregulates p53 and CDK inhibitor p21 protein levels, and downregulates Cdk4, Cdc2 and cyclin D2 protein levels (Ye and Zhang, 2004). The overexpression of heme oxygenase-1 (HO-1) in human pulmonary epithelial cells results in cell growth arrest during G0/G1 phase and increased resistance to hyperoxia (Lee et al., 1996).
RNR and cell cycle
RNR expression and activity are strictly regulated during cell cycle to generate and maintain proper dNTP pools that ensure the fidelity of DNA synthesis and repair (Tanaka et al., 2000; Sanvisens et al., 2011). Generally, the dNTP pools increase by 5- to 10-fold when cells transit from G0/G1 phase to S phase; this transition is mainly achieved by increased RNR levels during the exponential phase in bacteria and the S phase in eukaryotes (Cendra Mdel et al., 2013; Zhang et al., 2014a, b). In mammalian cells, the transcriptions of RRM1 and RRM2 are dependent on cell cycle with low or undetectable transcription levels in G0/G1 phase but maximal levels during S phase (Bjorklund et al., 1990; Chabes et al., 2004). In plants, the expressions of R1 and R2 are also S phase specific and dependent on the E2F-like motifs in their promoters (Chaboute et al., 2002). In budding yeast, the transcriptions of RNR genes (RNR1, RNR2 and RNR4) peak at the beginning of the S phase by regulating the transcription factor pairs Mbp1/Swi6 and Swi4/Swi6, which can bind to the Mlul cell cycle box (Sanvisens et al., 2013). However, no cell cycle regulation has been observed in RNR3 transcription (Lee and Elledge, 2006; Sanvisens et al., 2013).
Summary
Although results have indicated that iron-requiring proteins are implicated in DNA replication, repair and cell cycle control, limited information is available regarding their functional mechanisms. Several iron-requiring proteins, such as DNA polymerases/primases, DNA helicases, and RNRs, directly participate in DNA replication and repair. Biochemical and structural studies have suggested that Fe-S domains in these enzymes serve a structural rather than a redox-active function by possibly stabilizing local domain conformation that may mediate protein-protein or protein-nucleic acid interactions. Disruption of some iron-requiring proteins, particularly hemoproteins, associates with the generation of ROS, which results in DNA damage. Mutations of iron-requiring proteins are associated with diseases characterized by DNA repair defects and/or a poor response to replication stress in mammals. Thus, a detailed understanding of the mechanisms of Fe-requiring protein functions may provide insights into the related mutagenic diseases.
References
Acharya N, Johnson RE, Prakash S, Prakash L (2006) Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol 26:9555–9563
Alvaro D, Lisby M, Rothstein R (2007) Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet 3:e228
Amillet JM, Galiazzo F, Labbe-Bois R (1996) Effect of heme and vacuole deficiency on FRE1 gene expression and ferrireductase activity in Saccharomyces cerevisiae. FEMS Microbiol Lett 137:25–29
Anderson GJ, Vulpe CD (2009) Mammalian iron transport. Cellular Mol Life Sci CMLS 66:3241–3261
Andrews NC, Schmidt PJ (2007) Iron homeostasis. Annu Rev Physiol 69:69–85
Aslan M, Horoz M, Kocyigit A, Ozgonul S, Celik H, Celik M, Erel O (2006) Lymphocyte DNA damage and oxidative stress in patients with iron deficiency anemia. Mutat Res 601:144–149
Beaumont C (2010) Multiple regulatory mechanisms act in concert to control ferroportin expression and heme iron recycling by macrophages. Haematologica 95:1233–1236
Berthelet S, Usher J, Shulist K, Hamza A, Maltez N, Johnston A, Fong Y, Harris LJ, Baetz K (2010) Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcription factor Aft1 reveals iron-independent functions. Genetics 185:1111–1128
Bjorklund S, Skog S, Tribukait B, Thelander L (1990) S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry 29:5452–5458
Brown KR, Brown BM, Hoagland E, Mayne CL, Hegg EL (2004) Heme A synthase does not incorporate molecular oxygen into the formyl group of heme A. Biochemistry 43:8616–8624
Cendra Mdel M, Juarez A, Madrid C, Torrents E (2013) H-NS is a novel transcriptional modulator of the ribonucleotide reductase genes in Escherichia coli. J Bacteriol 195:4255–4263
Chabes AL, Pfleger CM, Kirschner MW, Thelander L (2003) Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA 100:3925–3929
Chabes AL, Bjorklund S, Thelander L (2004) S Phase-specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F-binding site and an upstream promoter activating region. J Biol Chem 279:10796–10807
Chaboute ME, Clement B, Philipps G (2002) S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5’ leader sequence. J Biol Chem 277:17845–17851
Chen KC, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson JJ (2000) Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell 11:369–391
Cheng NH, Zhang W, Chen WQ, Jin J, Cui X, Butte NF, Chan L, Hirschi KD (2011) A mammalian monothiol glutaredoxin, Grx3, is critical for cell cycle progression during embryogenesis. FEBS J 278:2525–2539
Coin F, Oksenych V, Egly JM (2007) Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 26:245–256
Couturier J, Touraine B, Briat JF, Gaymard F, Rouhier N (2013) The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions. Front Plant Sci 4:259
D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M (2012) Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149:1023–1034
Danilova N, Bibikova E, Covey TM, Nathanson D, Dimitrova E, Konto Y, Lindgren A, Glader B, Radu CG, Sakamoto KM et al. (2014). The role of DNA damage response in zebrafish and cellular models of Diamond Blackfan Anemia. Disease models & mechanisms
Deans AJ, West SC (2009) FANCM connects the genome instability disorders Bloom’s syndrome and Fanconi anemia. Mol Cell 36:943–953
Denic S, Agarwal MM (2007) Nutritional iron deficiency: an evolutionary perspective. Nutrition 23:603–614
Diaz-Castro J, Alferez MJ, Lopez-Aliaga I, Nestares T, Granados S, Barrionuevo M, Campos MS (2008) Influence of nutritional iron deficiency anemia on DNA stability and lipid peroxidation in rats. Nutrition 24:1167–1173
Dlouhy AC, Outten CE (2013) The iron metallome in eukaryotic organisms. Metal Ions Life Sci 12:241–278
Dunn LL, Suryo Rahmanto Y, Richardson DR (2007) Iron uptake and metabolism in the new millennium. Trends Cell Biol 17:93–100
Emerson LR, Nau ME, Martin RK, Kyle DE, Vahey M, Wirth DF (2002) Relationship between chloroquine toxicity and iron acquisition in Saccharomyces cerevisiae. Antimicrob Agents Chemother 46:787–796
Fregoso M, Laine JP, Aguilar-Fuentes J, Mocquet V, Reynaud E, Coin F, Egly JM, Zurita M (2007) DNA repair and transcriptional deficiencies caused by mutations in the Drosophila p52 subunit of TFIIH generate developmental defects and chromosome fragility. Mol Cell Biol 27:3640–3650
Fu D, Richardson DR (2007) Iron chelation and regulation of the cell cycle: 2 mechanisms of posttranscriptional regulation of the universal cyclin-dependent kinase inhibitor p21CIP1/WAF1 by iron depletion. Blood 110:752–761
Gan L, von Moltke LL, Trepanier LA, Harmatz JS, Greenblatt DJ, Court MH (2009) Role of NADPH-cytochrome P450 reductase and cytochrome-b5/NADH-b5 reductase in variability of CYP3A activity in human liver microsomes. Drug Metab Dispos 37:90–96
Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ (2012) MMS19 links cytoplasmic iron–sulfur cluster assembly to DNA metabolism. Science 337:243–245
Girvan HM, Munro AW (2013) Heme sensor proteins. J Biol Chem 288:13194–13203
Gkouvatsos K, Papanikolaou G, Pantopoulos K (2012) Regulation of iron transport and the role of transferrin. Biochim Biophys Acta 1820:188–202
Hamza A, Baetz K (2012) Iron-responsive transcription factor Aft1 interacts with kinetochore protein Iml3 and promotes pericentromeric cohesin. J Biol Chem 287:4139–4147
Haro KJ, Sheth A, Scheinberg DA (2012) Dysregulation of IRP1-mediated iron metabolism causes gamma ray-specific radioresistance in leukemia cells. PLoS ONE 7:e48841
Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28:739–745
Haunhorst P, Hanschmann EM, Brautigam L, Stehling O, Hoffmann B, Muhlenhoff U, Lill R, Berndt C, Lillig CH (2013) Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol Biol Cell 24:1895–1903
He L, Wang H, Jin H, Guo C, Xie H, Yan K, Li X, Shen Q, Qiao T, Chen G et al (2009) CIAPIN1 inhibits the growth and proliferation of clear cell renal cell carcinoma. Cancer Lett 276:88–94
Heath JL, Weiss JM, Lavau CP, Wechsler DS (2013) Iron deprivation in cancer—potential therapeutic implications. Nutrients 5:2836–2859
Herbik A, Bolling C, Buckhout TJ (2002) The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol 130:2039–2048
Heymann P, Ernst JF, Winkelmann G (2000) Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol Lett 186:221–227
Holmes-Hampton GP, Jhurry ND, McCormick SP, Lindahl PA (2013) Iron content of Saccharomyces cerevisiae cells grown under iron-deficient and iron-overload conditions. Biochemistry 52:105–114
Jiang X, Wang X (2004) Cytochrome C-mediated apoptosis. Annu Rev Biochem 73:87–106
Kamei A, Watanabe Y, Ishijima T, Uehara M, Arai S, Kato H, Nakai Y, Abe K (2010) Dietary iron-deficient anemia induces a variety of metabolic changes and even apoptosis in rat liver: a DNA microarray study. Physiol Genomics 42:149–156
Kaplan CD, Kaplan J (2009) Iron acquisition and transcriptional regulation. Chem Rev 109:4536–4552
Kaplan J, McVey Ward D, Crisp RJ, Philpott CC (2006) Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta 1763:646–651
Keyes SR, Cinti DL (1980) Biochemical properties of cytochrome b5-dependent microsomal fatty acid elongation and identification of products. J Biol Chem 255:11357–11364
Kilkenny ML, Longo MA, Perera RL, Pellegrini L (2013) Structures of human primase reveal design of nucleotide elongation site and mode of Pol alpha tethering. Proc Natl Acad Sci USA 110:15961–15966
Kumar D, Viberg J, Nilsson AK, Chabes A (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38:3975–3983
Laha S, Das SP, Hajra S, Sanyal K, Sinha P (2011) Functional characterization of the Saccharomyces cerevisiae protein Chl1 reveals the role of sister chromatid cohesion in the maintenance of spindle length during S-phase arrest. BMC Genet 12:83
Larade K, Jiang Z, Zhang Y, Wang W, Bonner-Weir S, Zhu H, Bunn HF (2008) Loss of Ncb5or results in impaired fatty acid desaturation, lipoatrophy, and diabetes. J Biol Chem 283:29285–29291
Lee YD, Elledge SJ (2006) Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev 20:334–344
Lee PJ, Alam J, Wiegand GW, Choi AM (1996) Overexpression of heme oxygenase-1 in human pulmonary epithelial cells results in cell growth arrest and increased resistance to hyperoxia. Proc Natl Acad Sci USA 93:10393–10398
Lesuisse E, Simon-Casteras M, Labbe P (1998) Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144(Pt 12):3455–3462
Li H, Outten CE (2012) Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry 51:4377–4389
Li L, Chen OS, McVey Ward D, Kaplan J (2001) CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276:29515–29519
Li L, Bagley D, Ward DM, Kaplan J (2008) Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol 28:1326–1337
Li L, Kaplan J (1998) Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem 273:22181–22187
Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE (2009) The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48:9569–9581
Lill R, Muhlenhoff U (2008) Maturation of iron–sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem 77:669–700
Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U (2012) The role of mitochondria in cellular iron–sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta 1823:1491–1508
Lipinski P, Starzynski RR, Drapier JC, Bouton C, Bartlomiejczyk T, Sochanowicz B, Smuda E, Gajkowska A, Kruszewski M (2005) Induction of iron regulatory protein 1 RNA-binding activity by nitric oxide is associated with a concomitant increase in the labile iron pool: implications for DNA damage. Biochem Biophys Res Commun 327:349–355
Lopez-Millan AF, Grusak MA, Abadia A, Abadia J (2013) Iron deficiency in plants: an insight from proteomic approaches. Front Plant Sci 4:254
Martinez-Pastor MT, de Llanos R, Romero AM, Puig S (2013) Post-transcriptional regulation of iron homeostasis in Saccharomyces cerevisiae. Int J Mol Sci 14:15785–15809
Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62:1191–1243
Miller JL (2013) Iron deficiency anemia: a common and curable disease. Cold Spring Harbor perspectives in medicine 3
Miyabe I, Kunkel TA, Carr AM (2011) The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet 7:e1002407
Muckenthaler MU, Galy B, Hentze MW (2008) Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr 28:197–213
Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E et al (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron–sulfur cluster. Cell Metab 12:373–385
Nakano K, Balint E, Ashcroft M, Vousden KH (2000) A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 19:4283–4289
Nasmyth K (1993) Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol 5:166–179
Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R (2010) Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat Chem Biol 6:758–765
Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ (2012) Eukaryotic DNA polymerases require an iron–sulfur cluster for the formation of active complexes. Nat Chem Biol 8:125–132
Ofir A, Kornitzer D (2010) Candida albicans cyclin Clb4 carries S-phase cyclin activity. Eukaryot Cell 9:1311–1319
Orrenius S, Nicotera P, Zhivotovsky B (2011) Cell death mechanisms and their implications in toxicology. Toxicol Sci Off J Soc Toxicol 119:3–19
Pamplona A, Ferreira A, Balla J, Jeney V, Balla G, Epiphanio S, Chora A, Rodrigues CD, Gregoire IP, Cunha-Rodrigues M et al (2007) Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria. Nat Med 13:703–710
Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L (2012) Mechanisms of mammalian iron homeostasis. Biochemistry 51:5705–5724
Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ (2006) The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci 119:4857–4865
Philpott CC (2006) Iron uptake in fungi: a system for every source. Biochim Biophys Acta 1763:636–645
Philpott CC, Rashford J, Yamaguchi-Iwai Y, Rouault TA, Dancis A, Klausner RD (1998) Cell-cycle arrest and inhibition of G1 cyclin translation by iron in AFT1-1(up) yeast. EMBO J 17:5026–5036
Philpott CC, Leidgens S, Frey AG (2012) Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta 1823:1509–1520
Poor CB, Wegner SV, Li H, Dlouhy AC, Schuermann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C (2014) Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci USA 111:4043–4048
Portnoy ME, Liu XF, Culotta VC (2000) Saccharomyces cerevisiae expresses three functionally distinct homologues of the nramp family of metal transporters. Mol Cell Biol 20:7893–7902
Portnoy ME, Jensen LT, Culotta VC (2002) The distinct methods by which manganese and iron regulate the Nramp transporters in yeast. Biochem J 362:119–124
Prakash S, Prakash L (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev 16:1872–1883
Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IK, Spies M (2008) The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J Biol Chem 283:1732–1743
Pyrih J, Harant K, Martincova E, Sutak R, Lesuisse E, Hrdy I, Tachezy J (2014) Giardia intestinalis incorporates heme into cytosolic cytochrome b(5). Eukaryot Cell 13:231–239
Quincozes-Santos A, Bobermin LD, Latini A, Wajner M, Souza DO, Goncalves CA, Gottfried C (2013) Resveratrol protects C6 astrocyte cell line against hydrogen peroxide-induced oxidative stress through heme oxygenase 1. PLoS ONE 8:e64372
Rao VA (2013) Iron chelators with topoisomerase-inhibitory activity and their anticancer applications. Antioxid Redox Signal 18:930–955
Rae TD, Goff HM (1998) The heme prosthetic group of lactoperoxidase. Structural characteristics of heme l and heme l-peptides. J Biol Chem 273:27968–27977
Reddy VV, Kupfer D, Caspi E (1977) Mechanism of C-5 double bond introduction in the biosynthesis of cholesterol by rat liver microsomes. J Biol Chem 252:2797–2801
Reid EL, Weynberg KD, Love J, Isupov MN, Littlechild JA, Wilson WH, Kelly SL, Lamb DC, Allen MJ (2013) Functional and structural characterisation of a viral cytochrome b5. FEBS Lett 587:3633–3639
Renton FJ, Jeitner TM (1996) Cell cycle-dependent inhibition of the proliferation of human neural tumor cell lines by iron chelators. Biochem Pharmacol 51:1553–1561
Romero A, Ramos E, de Los Rios C, Egea J, Del Pino J, Reiter RJ (2014) A review of metal-catalyzed molecular damage: protection by melatonin. J Pineal Res 56:343
Rouault TA (2012) Biogenesis of iron–sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Models Mech 5:155–164
Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF (2006) The DNA repair helicases XPD and FancJ have essential iron–sulfur domains. Mol Cell 23:801–808
Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR (2001) A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci USA 98:14322–14327
Sanvisens N, Bano MC, Huang M, Puig S (2011) Regulation of ribonucleotide reductase in response to iron deficiency. Mol Cell 44:759–769
Sanvisens N, de Llanos R, Puig S (2013) Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress, and iron bioavailability. Biomed J 36:51–58
Sauguet L, Klinge S, Perera RL, Maman JD, Pellegrini L (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS ONE 5:e10083
Schenkman JB, Jansson I (2003) The many roles of cytochrome b5. Pharmacol Ther 97:139–152
Schumacher SB, Stucki M, Hubscher U (2000) The N-terminal region of DNA polymerase delta catalytic subunit is necessary for holoenzyme function. Nucleic Acids Res 28:620–625
Severance S, Hamza I (2009) Trafficking of heme and porphyrins in metazoa. Chem Rev 109:4596–4616
Shah R, Agarwal AK (2013) Anemia associated with chronic heart failure: current concepts. Clin Interv Aging 8:111–122
Shakoury-Elizeh M, Protchenko O, Berger A, Cox J, Gable K, Dunn TM, Prinz WA, Bard M, Philpott CC (2010) Metabolic response to iron deficiency in Saccharomyces cerevisiae. J Biol Chem 285:14823–14833
Sharp P, Srai SK (2007) Molecular mechanisms involved in intestinal iron absorption. World J Gastroenterol 13:4716–4724
Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G (2002) Maturation of cytosolic iron–sulfur proteins requires glutathione. J Biol Chem 277:26944–26949
Siriwardana G, Seligman PA (2013) Two cell cycle blocks caused by iron chelation of neuroblastoma cells: separating cell cycle events associated with each block. Physiol Rep 1:e00176
Soe-Lin S, Apte SS, Andriopoulos B Jr, Andrews MC, Schranzhofer M, Kahawita T, Garcia-Santos D, Ponka P (2009) Nramp1 promotes efficient macrophage recycling of iron following erythrophagocytosis in vivo. Proc Natl Acad Sci USA 106:5960–5965
Solti A, Gaspar L, Meszaros I, Szigeti Z, Levai L, Sarvari E (2008) Impact of iron supply on the kinetics of recovery of photosynthesis in Cd-stressed poplar (Populus glauca). Ann Bot 102:771–782
Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9:3273–3297
Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R (2012) MMS19 assembles iron–sulfur proteins required for DNA metabolism and genomic integrity. Science 337:195–199
Stehling O, Mascarenhas J, Vashisht AA, Sheftel AD, Niggemeyer B, Rosser R, Pierik AJ, Wohlschlegel JA, Lill R (2013) Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron–sulfur proteins. Cell Metab 18:187–198
Sung P, Guzder SN, Prakash L, Prakash S (1996) Reconstitution of TFIIH and requirement of its DNA helicase subunits, Rad3 and Rad25, in the incision step of nucleotide excision repair. J Biol Chem 271:10821–10826
Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y (2000) A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404:42–49
Turrens JF (2003) Mitochondrial formation of reactive oxygen species. J Physiol 552:335–344
Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y (2007) Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol Biol Cell 18:2980–2990
Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y (2012) Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol Cell Biol 32:4998–5008
Urbanowski JL, Piper RC (1999) The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J Biol Chem 274:38061–38070
Vergeres G, Waskell L (1995) Cytochrome b5, its functions, structure and membrane topology. Biochimie 77:604–620
Vohradsky J (2012) Stochastic simulation for the inference of transcriptional control network of yeast cyclins genes. Nucleic Acids Res 40:7096–7103
Wagener FA, van Beurden HE, von den Hoff JW, Adema GJ, Figdor CG (2003) The heme–heme oxygenase system: a molecular switch in wound healing. Blood 102:521–528
Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE (2004) Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol 24:6891–6899
White MF, Dillingham MS (2012) Iron-sulphur clusters in nucleic acid processing enzymes. Curr Opin Struct Biol 22:94–100
Wu Y, Brosh RM Jr (2012) DNA helicase and helicase-nuclease enzymes with a conserved iron–sulfur cluster. Nucleic Acids Res 40:4247–4260
Wu H, Li L, Du J, Yuan Y, Cheng X, Ling HQ (2005) Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol 46:1505–1514
Wu Y, Suhasini AN, Brosh RM Jr (2009) Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell Mol Life Sci 66:1209–1222
Yamaguchi-Iwai Y, Ueta R, Fukunaka A, Sasaki R (2002) Subcellular localization of Aft1 transcription factor responds to iron status in Saccharomyces cerevisiae. J Biol Chem 277:18914–18918
Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, Mury SP, Capelluto DG, Finkielstein CV (2008) A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol Cell Biol 28:4697–4711
Ye H, Rouault TA (2010) Human iron–sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry 49:4945–4956
Ye W, Zhang L (2004) Heme controls the expression of cell cycle regulators and cell growth in HeLa cells. Biochem Biophys Res Commun 315:546–554
Yu B, Lane ME, Pestell RG, Albanese C, Wadler S (2000) Downregulation of cyclin D1 alters cdk 4- and cdk 2-specific phosphorylation of retinoblastoma protein. Mol Cell Biol Res Commun 3:352–359
Yu Y, Kovacevic Z, Richardson DR (2007) Tuning cell cycle regulation with an iron key. Cell Cycle 6:1982–1994
Yun CW, Ferea T, Rashford J, Ardon O, Brown PO, Botstein D, Kaplan J, Philpott CC (2000a) Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J Biol Chem 275:10709–10715
Yun CW, Tiedeman JS, Moore RE, Philpott CC (2000b) Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J Biol Chem 275:16354–16359
Yun CW, Bauler M, Moore RE, Klebba PE, Philpott CC (2001) The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J Biol Chem 276:10218–10223
Zhang AS (2010) Control of systemic iron homeostasis by the hemojuvelin-hepcidin axis. Adv Nutr 1:38–45
Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee DW, Bi E, Ohnishi T, Daldal F, Pain D et al (2008) Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol 28:5569–5582
Zhang F, Tao Y, Zhang Z, Guo X, An P, Shen Y, Wu Q, Yu Y, Wang F (2012) Metalloreductase Steap3 coordinates the regulation of iron homeostasis and inflammatory responses. Haematologica 97:1826–1835
Zhang C, Liu G, Huang M (2014a) Ribonucleotide reductase metallocofactor: assembly, maintenance and inhibition. Front Biol 9:104–113
Zhang Y, Li H, Zhang C, An X, Liu L, Stubbe J, Huang M (2014) Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America
Zhao N, Gao J, Enns CA, Knutson MD (2010) ZRT/IRT-like protein 14 (ZIP14) promotes the cellular assimilation of iron from transferrin. J Biol Chem 285:32141–32150
Acknowledgments
I would like to express my gratitude to Dr. Jianbin Wang, Dr. Guoqi Liu, and Dr. Weipeng Xiong for critically reading the manuscript and for facilitating discussions.
Compliance with ethics guidelines
Caiguo Zhang declares no conflict of interest.
This article does not contain any studies with human or animal as subjects performed by the author.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Zhang, C. Essential functions of iron-requiring proteins in DNA replication, repair and cell cycle control. Protein Cell 5, 750–760 (2014). https://doi.org/10.1007/s13238-014-0083-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-014-0083-7