Abstract
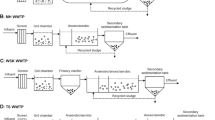
Liquid industrial chemical wastes are often treated with complex and integrated chemical and biological processes to attain the required standard for water discharge into the environment. Such wastewaters are potentially dangerous and of unknown composition. Biological treatment represents the cheapest means of pollution abatement. In the present work, a microbiological and genetic analysis was performed for the biological pool of an industrial wastewater treatment plant located in Civita Castellana (Viterbo, Italy). This study focused on active sludge treatment and on the subsequent secondary sedimentation system. The parameters of the biological section (the sludge active plant) are normally constantly monitored, but the biological efficiency of the microbial community should be better evaluated to understand the dynamics of the microbial community and its relationship to the overall standard quality parameters that are usually monitored. For this study, Biolog community level physiological profiling (CLPP) on EcoPlates and PCR-amplified 16S rRNA denaturing gradient gel electrophoresis (DGGE) were used in comparison and combined as ecological techniques to characterize an anthropic closed ecosystem. Biolog CLPP provides the potential metabolic pattern and DGGE analyses helps to explain the structure and complexity of the microbial community. The results suggest that these techniques could be predictive and more useful when used together than alone. In addition, a principal component analysis (PCA) performed on the Biolog assay over time can relate principal components predictive wells to the biological and standard chemical analysis used to control and monitor the activity of this type of industrial wastewater treatment plant, providing a selection method for further DGGE community analysis.





Similar content being viewed by others

References
El Fantroussi S, Verschuere L, Verstraete W, Top EM (1999) Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol 65(3):982–988
Fischer SG, Lerman LS (1980) Separation of random fragments of DNA according to properties of their sequences. Proc Natl Acad Sci USA 77(8):4420–4424
Frąc M, Oszust K, Lipiec J (2012) Community Level Physiological Profiles (CLPP), characterization and microbial activity of soil amended with dairy sewage sludge. Sensors 12:3253–3268. doi:10.3390/s120303253
Garland J (1997) Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol Ecol 24:289–300
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359. doi:10.1007/978[-‐]1-60761-439-5_16
Green SJ, Leigh MB, Neufeld JD (2009) Denaturing gradient gel electrophoresis (DGGE) for microbial community analysis. In: Timmis KN (ed) Microbiology of hydrocarbons, oils, lipids, and derived compounds. Springer, Heidelberg, pp 4137–4158
Harch BD, Correll RL, Meech W, Kirkby CA, Pankhurst CE (1997) Using the Gini coefficient with BIOLOG substrate utilization data to provide an alternative quantitative measure for comparing bacterial soil communities. J Microbiol Methods 30:91–101
Hitzl W, Rangger A, Sharma S, Insam H (1996) Separation power of the 95 substrates of the BIOLOG system determined in various soils. FEMS Microbiol Ecol 22:167–174. doi:10.1111/j.1574-6941.1997.tb00368.x
İnceoğlu Ö, Falcão Salles J, Van Elsas JD (2012) Soil and cultivar type shape the bacterial community in the potato rhizosphere. Microb Ecol 63(2):460–470. doi:10.1007/s00248-011-9930-8
Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W (2008) How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol 10:1571–1581
Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59(3):695–700
Myers RM, Fisher SG, Lerman LS, Maniatis T (1985) Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res 13(9):3131–3145
Preston-Mafham J, Boddy L, Randerson PF (2002) Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles—a critique. FEMS Microbiol Ecol 42:1–14
Schmitt H, Van Beelen P, Tolls J, Van Leeuwen CL (2004) Pollution-induced community tolerance of soil microbial communities caused by the antibiotic sulfachloropyridazine. Environ Sci Technol 38(4):1148–1153. doi:10.1021/es034685p
Shi W, Bischoff M, Turco R, Konopka A (2005) Microbial catabolic diversity in soils contaminated with hydrocarbons and heavy metals. Environ Sci Technol 39(7):1974–1979
Shishido M, Chanway CP (1998) Forest soil community responses to plant growth-promoting rhizobacteria and spruce seedlings. Biol Fertil Soils 26(3):179–186
Smalla K, Wachtendorf U, Heuer H, Liu WT, Forney L (1998) Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl Environ Microbiol 64(4):1220–1225
Song C, Yang C, Liu R, Li Q, Li B, Wang S, Mulchandani A (2011) Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ Sci Technol 45(17):7408–7415. doi:10.1021/es2010545
Stefanowicz A (2006) The Biolog plates technique as a tool in ecological studies of microbial communities. Pol J Environ Stud 15(5):669–676, and references therein
Travis ER, Hannink NK, Van der Gast CJ, Thompson IP, Rosser SJ, Bruce NC (2007) Impact of transgenic tobacco on trinitrotoluene (TNT) contaminated soil community. Environ Sci Technol 41(16):5854–5861. doi:10.1021/es070507a
Valášková V, Baldrian P (2009) Denaturing gradient gel electrophoresis as a fingerprinting method for the analysis of soil microbial communities. Plant Soil Environ 55(10):413–423
Victorio L, Gilbride KA, Grant Allen D, Liss SN (1996) Phenotypic fingerprinting of microbial communities in wastewater treatment systems. Water Res 30(5):1077–1086
Ward DM, Bateson MM, Weller R, Ruff-Roberts AL (1992) Ribosomal RNA analysis of microorganism as they occur in nature. Adv Microbial Ecol 12:219–286. doi:10.1145/267521.267546
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 37:463–464. doi:10.1099/00207713[-‐]37-4-463
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Moretti, G., Matteucci, F., Ercole, C. et al. Microbial community distribution and genetic analysis in a sludge active treatment for a complex industrial wastewater: a study using microbiological and molecular analysis and principal component analysis. Ann Microbiol 66, 397–405 (2016). https://doi.org/10.1007/s13213-015-1122-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1122-1