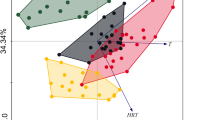
Abstract—The microbial diversity and community structure of activated sludge in the biological treatment units of five municipal WWTPs and two industrial WWTPs were studied. The results showed that industrial WWTPs had the lowest diversity and abundance of activated sludge microbial communities, indicating that industrial wastewater inhibited the growth of microbial communities. In the correlation analysis between environmental factors and microbial diversity indices, pH and COD showed a significant negative correlation with the number of OTUs and microbial diversity. The main dominant microbial genus were Hyphomicrobium, Acinetobacter, Novosphingobium, Acinetobacter, and Meiothermus. Both principal coordinate analysis (PCoA) and phylogenetic analysis showed large differences in the structure of municipal and industrial microbial community structure, indicating the uniqueness of the activated sludge community structure in industrial WWTPs. Gene function predictions indicated that LJ had higher metabolic function associated with inorganic salt and lipid metabolism. Further co-occurrence network analysis showed that many of the key industrial WWTPs belonged in the same module, suggesting that they had strong interspecific interactions but were relatively independent from other microorganisms in the municipal WWTPs. This study broadens our understanding of microbial community differences between municipal WWTPs and industrial WWTPs.


Similar content being viewed by others
REFERENCES
Banerjee, S., Schlaeppi, K., and van der Heijden, M.G., Keystone taxa as drivers of microbiome structure and functioning, Nat. Rev. Microbiol., 2018, vol. 16, no. 9, pp. 567–576.
Cao, J., Zhang, T., Wu, Y., Sun, Y., Zhang, Y., Huang, B., Fu, B., Yang, E., Zhang, Q., and Luo, J., Correlations of nitrogen removal and core functional genera in full-scale wastewater treatment plants: influences of different treatment processes and influent characteristics, Bioresour. Technol., 2020, vol. 297, p. 122455.
Chen, Y., Guo, X., Feng, J., Lu, D., Niu, Z., Tou, F., Hou, L., Liu, M., and Yang, Y., Impact of ZnO nanoparticles on the antibiotic resistance genes (ARGs) in estuarine water: ARG variations and their association with the microbial community, Environ. Sci.: Nano, 2019, vol. 6, no. 8, pp. 2405–2419.
Czarny, J., Staninska-Pięta, J., Piotrowska-Cyplik, A., Juzwa, W., Wolniewicz, A., and Marecik, R., Acinetobacter sp. as the key player in diesel oil degrading community exposed to PAHs and heavy metals, J. Hazard Mater., 2020, vol. 383, p. 121168.
Dong, X., He, Y., Peng, X., and Jia, X., Triclosan in contact with activated sludge and its impact on phosphate removal and microbial community, Bioresour. Technol., 2021, vol. 319, p. 124134.
Ekman, J., Kosonen, M., Jokela, S., Kolari, M., Korhonen, P., and Salkinoja-Salonen, M., Detection and quantitation of colored deposit-forming Meiothermus spp. in paper industry processes and end products, J. Ind. Microbiol. Biotechnol., 2007, vol. 34, no. 3, pp. 203–211.
Faust, K. and Raes, J., Microbial interactions: from networks to models, Nat. Rev. Microbiol., 2012, vol. 10, no. 8, pp. 538–550.
Gao, P., Xu, W., Sontag, P., Li, X., Xue, G., Liu, T., and Sun, W., Correlating microbial community compositions with environmental factors in activated sludge from four full-scale municipal wastewater treatment plants in Shanghai, China, Appl. Microbiol. Biotechnol., 2016, vol. 100, no. 10, pp. 4663–4673.
Guo, J., Ni, B., Han, X., Chen, X., Bond, P., Peng, Y., and Yuan, Z., Unraveling microbial structure and diversity of activated sludge in a full-scale simultaneous nitrogen and phosphorus removal plant using metagenomic sequencing, Enzyme Microbiol. Technol., 2017, vol. 102, pp. 16–25.
Hall, E. R., Monti, A., and Mohn, W. W., A comparison of bacterial populations in enhanced biological phosphorus removal processes using membrane filtration or gravity sedimentation for solids–liquid separation, Water Res., 2010, vol. 44, no. 9, pp. 2703–2714.
Hreiz, R., Latifi, M. A., and Roche, N., Optimal design and operation of activated sludge processes: state-of-the-art, Chem. Eng. J., 2015, vol. 281, pp. 900–920.
Huang, M., Li, Y., and Gu, G., Chemical composition of organic matters in domestic wastewater, Desalination., 2010, vol. 262, nos. 1–3, pp. 36–42.
Ibarbalz, F. M., Figuerola, E. L., and Erijman, L., Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks, Water Res., 2013, vol. 47, no. 11, pp. 3854–3864.
Jiang, Y., Li, R., Yang, Y., Yu, M., Xi, B., Li, M., Xu, Z., Gao, S., and Yang, C., Migration and evolution of dissolved organic matter in landfill leachate-contaminated groundwater plume, Resour., Conserv. Recycl., 2019, vol. 151, p. 104463. Kamika, I., Coetzee, M., Mamba, B. B., Msagati, T., and Momba, M. N. B., The Impact of Microbial Ecology and Chemical Profile on the Enhanced Biological Phosphorus Removal (EBPR) Process: A Case Study of Northern Wastewater Treatment Works, Johannesburg, International Journal of Environmental Research and Public Health., 2014, vol. 11, no. 3, pp. 2876–2898.
Kim, E., Yulisa, A., Kim, S., and Hwang, S., Monitoring microbial community structure and variations in a full-scale petroleum refinery wastewater treatment plant, Bioresour. Technol., 2020, vol. 306, p. 123178.
Kristiansen, R., Nguyen, H.T.T., Saunders, A. M., Nielsen, J.L., Wimmer, R., Le, V.Q., McIlroy, S.J., Petrovski, S., Seviour, R J., Calteau, A., Nielsen, K.L., and Nielsen, P.H., A metabolic model for members of the genus Tetrasphaera involved in enhanced biological phosphorus removal, ISME J., 2013, vol. 7, no. 3, pp. 543–554.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K., MEGA X: molecular evolutionary genetics analysis across computing platforms, Mol. Biol. Evol., 2018, vol. 35, no. 6, pp. 1547–1549.
Li, Y., Wu, Y., Wang, S., and Jia, L., Effect of organic loading on phosphorus forms transformation and microbial community in continuous-flow A2/O process, Water Sci. Technol., 2021, vol. 83, no. 11, pp. 2640–2651.
Ou, D., Li, H., Li, W., Wu, X., Wang, Y., and Liu, Y., Salt-tolerance aerobic granular sludge: formation and microbial community characteristics, Bioresour. Technol., 2018a, vol. 249, pp. 132–138.
Ou, D., Li, W., Li, H., Wu, X., Li, C., Zhuge, Y., and Liu, Y., Enhancement of the removal and settling performance for aerobic granular sludge under hypersaline stress, Chemosphere., 2018b, vol. 212, pp. 400–407.
Silva-Bedoya, L.M., Sánchez-Pinzón, M.S., Cadavid-Restrepo, G.E., and Moreno-Herrera, C.X., Bacterial community analysis of an industrial wastewater treatment plant in Colombia with screening for lipid-degrading microorganisms, Microbiol. Res., 2016, vol. 192, pp. 313–325.
Stadler, L.B. and Love, N.G., Impact of microbial physiology and microbial community structure on pharmaceutical fate driven by dissolved oxygen concentration in nitrifying bioreactors, Water Res., 2016, vol. 104, pp. 189–199.
Wang, X., Xia, Y., Wen, X., Yang, Y., and Zhou, J., Microbial community functional structures in wastewater treatment plants as characterized by GeoChip, PLoS One, 2014a, vol. 9, no. 3, p. e93422.
Wang, Z., Zhang, X., Lu, X., Liu, B., Li, Y., Long, C., and Li, A., Abundance and diversity of bacterial nitrifiers and denitrifiers and their functional genes in tannery wastewater treatment plants revealed by high-throughput sequencing, PLoS One, 2014b, vol. 9, no. 11, p. e113603.
Xu, G., Zhang, W., and Xu, H., Can dispersions be used for discriminating water quality status in coastal ecosystems? A case study on biofilm-dwelling microbial eukaryotes, Ecol. Indic., 2015, vol. 57, pp. 208–214.
Xu, S., Yao, J., Ainiwaer, M., Hong, Y., and Zhang, Y., Analysis of bacterial community structure of activated sludge from wastewater treatment plants in winter, Biomed. Res. Int., 2018, vol. 2018, pp. 1–8.
Yang, Q., Xiong, P., Ding, P., Chu, L., and Wang, J., Treatment of petrochemical wastewater by microaerobic hydrolysis and anoxic/oxic processes and analysis of bacterial diversity, Bioresour. Technol., 2015, vol. 196, pp. 169–175.
Yang, Y., Wang, L., Xiang, F., Zhao, L., and Qiao, Z., Activated sludge microbial community and treatment performance of wastewater treatment plants in industrial and municipal zones, Int. J. Environ. Res. Public Health, 2020, vol. 17, no. 2, p. 436.
Yu, K., and Zhang, T., Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge, PLoS One., 2012, vol. 7, no. 5, p. e38183.
Yuan, J., Lai, Q., Zheng, T., and Shao, Z., Novosphingobium indicum sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from a deep-sea environment, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, no. 8, pp. 2084–2088.
Zhang, B., Xu, X., and Zhu, L., Structure and function of the microbial consortia of activated sludge in typical municipal wastewater treatment plants in winter, Sci. Rep. (UK), 2017, vol. 7, no. 1, pp. 1–11.
Zhang, Y., Ji, Z., and Pei, Y., Nutrient removal and microbial community structure in an artificial-natural coupled wetland system, Process Saf. Environ., 2021, vol. 147, p. 1160–1170.
Zhou, X., Zhang, J., and Wen, C., Community composition and abundance of anammox bacteria in cattail rhizosphere sediments at three phenological stages, Curr. Microbiol., 2017, vol. 74, no. 11, pp. 1349–1357.
Funding
This work was supported by the Project supported by the Science and Technology Research of Henan Province, China (222102320395), the Basic research plan for key scientific research projects of higher education institutions in Henan Province, China (20B610001, 21B610003), Training Program for Young Key Teachers of higher education institutions in Henan Province, China (2020GGJS218), Innovative training program for college students in Henan Province, China (S202111765048).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
The authors have no financial conflicts of interest to declare.
This article adheres to ethical standards. The authors are committed to ethical research guidelines, including compliance with laboratory safety regulations, respect for participants' privacy, and protection of participants' physical and psychological well-being, and the authors will ensure that all study results are truthful and free from any deception.
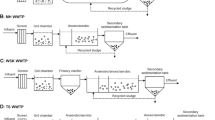
WASTEWATER TREATMENT PROCESS
In briefly, BF, BY and PDS were collected from municipal WWTPs which used the oxidation ditch process. YZA and YZB was collected from municipal WWTPs which used anaerobic-anoxic-oxic process. LJ was collected at the WWTP of a chlor-alkali chemical company, which mainly produces basic chemical raw materials such as caustic soda and PVC resin. The wastewater in LJ was first neutralized by acid-base, followed by hydrolytic acidification and contact oxidation. LLY was collected at the WWTP of a nylon chemical company, whose main products were caprolactam, cyclohexanol, and cyclohexanoic acid The LLY treated effluent was firstly pretreated with amoximation, then passed through the Bio-stability Bed and entered the anaerobic tank, followed by the MSBR (Modified Sequencing Batch Reactor).
Supplementary Information
Rights and permissions
About this article
Cite this article
Ziyan Yang, Guo, F., Wang, Q. et al. Diversity of Activated Sludge Microbial Community Structure in Different Wastewater Treatment Plants. Biol Bull Russ Acad Sci 50, 329–337 (2023). https://doi.org/10.1134/S1062359023700383
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1062359023700383