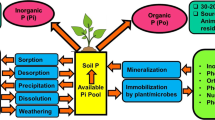
Abstract
Soil is a treasure chest for beneficial bacteria with applications in diverse fields, which include agriculture, rhizoremediation, and medicine. Metagenomic analysis of four soil samples identified Proteobacteria as the dominant phylum (32–52%) followed by the phylum Acidobacteria (11–21% in three out of four soils). Bacteria that were prevalent at the highest level belong to the genus Kaistobacter (8–19%). PICRUSt analysis predicted KEGG functional pathways associated with the metagenomes of the four soils. The identified pathways could be attributed to metal tolerance, antibiotic resistance and plant growth promotion. The prevalence of phosphate solubilizing bacteria (PSB) was investigated in four soil samples, ranging from 26 to 59% of the total culturable bacteria. The abundance of salt-tolerant and metal-tolerant bacteria showed considerable variation ranging from 1 to 62% and 4–69%, respectively. In comparison, the soil with the maximum prevalence of temperature-tolerant and antibiotic-resistant bacteria was close 30%. In this study, the common pattern observed was that PSB were the most abundant in all types of soils compared to other traits. Conversely, most of the isolates, which are salt-tolerant, copper-tolerant, and ampicillin-resistant, showed phosphate solubilization activity. The sequencing of the partial 16S-rRNA gene revealed that PSB belonged to Bacillus genera.








Similar content being viewed by others
Abbreviations
- PSB:
-
Phosphate solubilizing bacteria
- STB:
-
Salt-tolerant bacteria
- MTB:
-
Metal-tolerant bacteria
- ARB:
-
Antibiotic-resistant bacteria
- PGPB:
-
Plant growth-promoting bacteria
- ICP-MS:
-
Inductively coupled plasma mass spectrometry
- NGS:
-
Next-generation sequencing
- MSA:
-
Multiple sequence alignment
- BIC:
-
Bayesian information criterion
- ML:
-
Maximum-likelihood
- MRGs:
-
Metal resistance genes
- ARGs:
-
Antibiotic-resistance genes
References
Albdaiwi RN, Khaymi-Horani H, Ayad JY, Alananbeh KM, Kholoud M, Al-Sayaydeh R (2019) Isolation and characterization of halotolerant plant growth promoting rhizobacteria from durum wheat (Triticum turgidum subsp. durum) cultivated in saline areas of the dead sea region. Front Microbiol 10:1639
Alori ET, Glick BR, Babalola OO (2017) Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol 8:971
Altimira F, Yáñez C, Bravo G, González M, Rojas LA, Seeger M (2012) Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol 12:193
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Azizoglu U (2019) Bacillus thuringiensis as a biofertilizer and biostimulator: a mini-review of the little-known plant growth-promoting properties of Bt. Curr Microbiol 76:1379–1385
Bell JH, Thrun L, LeBeau M, Makarevitch I, Goldberg J, Martin P (2016) Antibiotic resistance genes detection in environmental samples. CourseSource. https://doi.org/10.24918/cs.2016.3
Cervantes C, Gutierrez-Corona F (1994) Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol Rev 14:121–137
Chaparro JM, Sheflin AM, Manter DK, Vivanco JM (2012) Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 48:489–499
Chen J, Li J, Zhang H, Shi W, Liu Y (2019) Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front Microbiol 10:1916
Choudhary DK, Kasotia A, Jain S, Vaishnav A, Kumari S, Sharma KP, Varma A (2016) Bacterial-mediated tolerance and resistance to plants under abiotic and biotic stresses. J Plant Growth Regul 35:276–300
Choudhary M, Sharma PC, Jat HS, Dash A, Rajashekar B, McDonald AJ, Jat ML (2018) Soil bacterial diversity under conservation agriculture-based cereal systems in Indo-Gangetic Plains. 3 Biotech 8:304
Classen AT, Sundqvist MK, Henning JA, Newman GS, Moore JA, Cregger MA, Moorhead LC, Patterson CM (2015) Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: what lies ahead? Ecosphere 6:1–21
Cota-Ruiz K, de Los L, Santos Y, Hernández-Viezcas JA, Delgado-Rios M, Peralta-Videa JR, Gardea-Torresdey JL (2019) A comparative metagenomic and spectroscopic analysis of soils from an international point of entry between the US and Mexico. Environ Int 123:558–566
Dogra N, Yadav R, Kaur M, Adhikary A, Kumar S, Ramakrishna W (2019) Nutrient enhancement of chickpea grown with plant growth promoting bacteria in local soil of Bathinda, Northwestern India. Physiol Mol Biol Plants 25:1251–1259
Douglas GM, Maffei VJ, Zaneveld JR et al (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688
Dube JP, Valverde A, Steyn JM, Cowan DA, van der Waals JE (2019) Differences in bacterial diversity, composition and function due to long-term agriculture in soils in the eastern free state of South Africa. Diversity 11:61
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Eida AA, Ziegler M, Lafi FF, Michell CT, Voolstra CR, Hirt H, Saad MM (2018) Desert plant bacteria reveal host influence and beneficial plant growth properties. PLoS ONE 13:e0208223
El Attar I, Taha K, El Bekkay B, El Khadir M, Alami IT, Aurag J (2019) Screening of stress tolerant bacterial strains possessing interesting multi-plant growth promoting traits isolated from root nodules of Phaseolus vulgaris L. Biocatal Agric Biotechnol 20:101225
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nature Rev Microbiol 15:579
Greene NP, Kaplan E, Crow A, Koronakis V (2018) Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front Microbiol 9:950
Gupta A, Joia J, Sood A, Sood R, Sidhu C, Kaur G (2016) Microbes as potential tool for remediation of heavy metals: a review. J Microb Biochem Technol 8:364–372
Hu HW, Wang JT, Li J, Li JJ, Ma YB, Chen D, He JZ (2016) Field-based evidence for copper contamination induced changes of antibiotic resistance in agricultural soils. Environ Microbiol 18:3896–3909
Johnson JS, Spakowicz DJ, Hong B, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM (2019) Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun 10:5029
Kang C-H, Kwon Y-J, So J-S (2016) Bioremediation of heavy metals by using bacterial mixtures. Ecol Engg 89:64–69
Karnwal A (2019) Screening, isolation and characterization of culturable stress-tolerant bacterial endophytes associated with Salicornia brachiata and their effect on wheat (Triticum aestivum L.) and maize (Zea mays) growth. J Plant Prot Res 59:293–303
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Langille M, Zaneveld J, Caporaso J et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Li K, Ramakrishna W (2011) Effect ofmultiple metal resistant bacteria from contaminated lake sediments on metal accumulation and plant growth. J Hazard Mater 189:531–539
Liu X, Zhang S, Jiang Q, Bai Y, Shen G, Li S, Ding W (2016) Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci Rep 6:36773
Mahanty T, Bhattacharjee S, Goswami M, Bhattacharyya P, Das B, Ghosh A, Tribedi P (2017) Biofertilizers: a potential approach for sustainable agriculture development. Env Sci Pollut Res 24:3315–3335
Marcel M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12
Moreno-Espíndola IP, Ferrara-Guerrero MJ, Luna-Guido ML, Ramírez-Villanueva DA, De León-Lorenzana AS, Gómez-Acata S, González-Terreros E, Ramírez-Barajas B, Navarro-Noya YE, Sánchez-Rodríguez LM, Fuentes-Ponce M, Macedas-Jímenez JU, Dendooven L (2018) The bacterial community structure and microbial activity in a traditional organic Milpa farming system under different soil moisture conditions. Front Microbiol 9:2737
NCBI Resource Coordinators (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44(D1):D7–D19
Nelkner J, Henke C, Lin TW, Pätzold W, Hassa J, Jaenicke S, Grosch R, Pühler A, Sczyrba A, Schlüter A (2019) Effect of long-term farming practices on agricultural soil microbiome members represented by metagenomically assembled genomes (MAGs) and their predicted plant-beneficial genes. Genes (basel) 10:24
Nyoyoko VF, Anyanwu CU (2019) Isolation and screening of heavy metal resistant ammonia oxidizing bacteria from soil and waste dump: a potential candidate for bioremediation of heavy metals. J Appl Microb Res 3:15–24
Ojuederie OB, Babalola OO (2017) Microbial and plant-assisted bioremediation of heavy metal polluted environments: a review. Int J Env Res Public Health 14:1504
Raftery AE (1999) Bayes factors and BIC: comment on “a critique of the Bayesian information criterion for model selection.” Soc Methods Res 27:411–427
Ramakrishna W, Yadav R, Li K (2019) Plant growth promoting bacteria in agriculture: two sides of a coin. Appl Soil Ecol 138:10–18
Ramakrishna W, Rathore P, Kumari R, Yadav R (2020) Brown gold of marginal soil: Plant growth promoting bacteria to overcome plant abiotic stress for agriculture, biofuels and carbon sequestration. Sci Total Environ 711:135062
Reinhold-Hurek B, Bünger W, Burbano CS, Sabale M, Hurek T (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu Rev Phytopathol 53:403–424
Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351
Santoyo G, Pacheco CH, Salmerón JH, León RH (2017) The role of abiotic factors modulating the plant-microbe-soil interactions: toward sustainable agriculture. A review. Spanish J Agric Res 15:13
Sá-Pereira P, Rodrigues M, Sim[otilde]es F, Domingues L, e Castro IV (2009) Bacterial activity in heavy metals polluted soils: metal efflux systems in native rhizobial strains. Geomicrobiol J 26(4):281–288
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501
Seiler C, Berendonk TU (2012) Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:399
Solanki MK, Wang Z, Wang F-Y, Li C-N, Gupta CL, Singh RK, Malviya MK, Singh P, Yang L-T, Li Y-R (2020) Assessment of diazotrophic proteobacteria in sugarcane rhizosphere when intercropped with legumes (peanut and soybean) in the field. Front Microbiol 11:1814
Sun L, Wang X, Li Y (2016) Increased plant growth and copper uptake of host and non-host plants by metal-resistant and plant growth-promoting endophytic bacteria. Int J Phytoremed 18:494–501
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thummeepak R, Pooalai R, Harrison C, Gannon L, Thanwisai A, Chantratita N, Millard AD, Sitthisak S (2020) Essential gene clusters involved in copper tolerance identified in Acinetobacter baumannii clinical and environmental isolates. Pathogens 9:60
Tirry N, Joutey NT, Sayel H, Kouchou A, Bahafid W, Asri M, El Ghachtouli N (2018) Screening of plant growth promoting traits in heavy metals resistant bacteria: prospects in phytoremediation. J Genet Engg Biotechnol 16:613–619
Tiwari S, Prasad V, Lata C (2019) Bacillus: plant growth promoting bacteria for sustainable agriculture and environment. In: Singh JS, Singh DP (eds) New and future developments in microbial biotechnology and bioengineering. Elsevier, pp 43–55
Torres MDR, Alcaraz LD, Souza V, Olmedo-Álvarez G (2018) Single genus approach to understanding bacterial diversity, niche, distribution, and genomics: the Bacillus in Cuatro Ciénegas. In: Souza V, Olmedo-Álvarez G, Eguiarte L (eds) Cuatro Ciénegas ecology, natural history and microbiology. Cuatro Ciénegas basin: an endangered hyperdiverse oasis. Springer, Cham
Verma JP, Jaiswal DK, Krishna R, Prakash S, Yadav J, Singh V (2018) Characterization and screening of thermophilic Bacillus strains for developing plant growth promoting consortium from hot spring of Leh and Ladakh region of India. Front Microbiol 9:1293
Wei YJ, Wu Y, Yan YZ, Zou W, Xue J, Ma WR, Wang W, Tian G, Wang LY (2018) High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS ONE 13:e0193097
Woodman ME, Savage CR, Arnold WK, Stevenson B (2016) Direct PCR of intact bacteria (colony PCR). Curr Prot Microbiol. https://doi.org/10.1002/cpmc.14
Wu S-H, Huang B-H, Huang C-L, Li G, Liao P-C (2018) The aboveground vegetation type and underground soil property mediate the divergence of soil microbiomes and the biological interactions. Microb Ecol 75:434–446
Yadav R, Ror P, Rathore P, Ramakrishna W (2020a) Bacteria from native soil in combination with arbuscular mycorrhizal fungi augment wheat yield and biofortification. Plant Physiol Biochem 150:222–233
Yadav R, Ror P, Rathore P, Kumar S, Ramakrishna W (2020b) Bacillus subtilis CP4, isolated from native soil in combination with arbuscular mycorrhizal fungi promotes biofortification, yield and metabolite production in wheat under field conditions. J Appl Microbiol. https://doi.org/10.1111/jam.14951
Yang R, Luo C, Chen Y, Wang G, Xu Y, Shen Z (2013) Copper-resistant bacteria enhance plant growth and copper phytoextraction. Int J Phytoremed 15:573–584
Yao A, Bochow H, Karimov S, Boturov U, Sanginboy S, Sharipov A (2006) Effect of FZB 24® Bacillus subtilis as a biofertilizer on cotton yields in field tests. Arch Phytopathol Plant Prot 39:323–328
Ye J, McGinnis S, Madden TL (2006) BLAST: improvements for better sequence analysis. Nucleic Acids Res 34:W6–W9
Zhang Y, Shen H, He X, Thomas BW, Lupwayi NZ, Hao X, Thomas MC, Shi X (2017) Fertilization shapes bacterial community structure by alteration of soil pH. Front Microbiol 8:1325
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
13205_2021_2904_MOESM1_ESM.pptx
Supplementary file1 (PPTX 829 KB) Fig. S1. Map showing areas in Punjab and Mizoram from where soil samples were collected. White colored dot in Punjab map is the site from where Ramsara soil was collected, blue colored dot is the site from where rice and cotton field samples were collected and white dot in Mizoram map is the site from where Aizawl sample was collected. Maps were downloaded from https://gramener.com/indiamap/
13205_2021_2904_MOESM3_ESM.pptx
Supplementary file3 (PPTX 84 KB) Fig. S3. Rarefaction plot for four samples of V3-V4 region at a depth of 2,00,000. The number of sequences per sample are shown on x-axis versus nember of observed species on y-axis. The samples have been colored by their respective names.
13205_2021_2904_MOESM4_ESM.pptx
Supplementary file4 (PPTX 538 KB) Fig. S4. Maximum likelihood (ML) phylogram based on 16S rRNA gene sequences using the TN92+G model of molecular evolution in MEGA X. Numbers near nodes represent the ML bootstrap proportion. Bootstrapping was performed with 1000 replicates. The sequences of soil bacteria that are part of the current study are in bold. This phylogram is rooted with Pseudomonas aeruginosa (LN874213) sequence as an outgroup. Scale bar is given on the bottom.
Rights and permissions
About this article
Cite this article
Rathore, P., Joy, S.S., Yadav, R. et al. Co-occurrence and patterns of phosphate solubilizing, salt and metal tolerant and antibiotic-resistant bacteria in diverse soils. 3 Biotech 11, 356 (2021). https://doi.org/10.1007/s13205-021-02904-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02904-7