Abstract
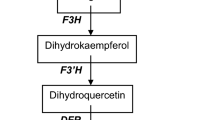
microRNAs (miRNAs) play critical regulatory roles in plant growth and development. In the present study, the function of herbaceous peony (Paeonia lactiflora Pall.) miR156e-3p in the regulation of color formation has been investigated. Firstly, P. lactiflora miR156e-3p precursor sequence (pre-miR156e-3p) was isolated. Subsequently, the overexpression vector of pre-miR156e-3p was constructed and transformed into Arabidopsis thaliana. Moreover, the medium screening, GUS staining, polymerase chain reaction (PCR) of the GUS region and real-time quantitative PCR (qRT-PCR) of miR156e-3p all confirmed that the purpose gene had been successfully transferred into Arabidopsis plants and expressed, which resulted in apparent purple lateral branches. And this change in color was caused by the improved anthocyanin accumulation. In addition, expression analysis had shown that the level of miR156e-3p transcript was increased, while transcription level of target gene squamosa promoter binding protein-like gene (SPL1), encoding SPL transcription factor that negatively regulated anthocyanin accumulation, was repressed in miR156e-3p-overexpressing transgenic plants, and its downstream gene dihydroflavonol 4-reductase gene (DFR) that was directly involved in anthocyanin biosynthesis was strongly expressed, which resulted in anthocyanin accumulation of Arabidopsis lateral branches. These findings would improve the understanding of miRNAs regulation of color formation in P. lactiflora.






Similar content being viewed by others
References
Chen Z, Gao X, Zhang J (2015) Alteration of osa-miR156e expression affects rice plant architecture and strigolactones (SLs) pathway. Plant Cell Rep 34:767–781
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2014) The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J 80:1108–1117
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23:1512–1522
He H, Liang G, Li Y, Wang F, Yu D (2014) Two young microRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol 164:853–865
Hu G, Fan J, Xian Z, Huang W, Lin D, Li Z (2014) Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci 229:86–95
Jia XY, Yu ZQ, Liang JP, Tang GL, Jin LH, Zhang L, He LH, Li RZ (2013) Cloning of Arabidopsis At-pri-miR828 gene and its genetic transformation into tomato. Acta Hortic Sin 40:2419–2428 (in Chinese)
Jian XY, Zhang L, Li GL, Zhang L, Wang XJ, Cao XF, Fang X, Chen F (2010) Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics 95:47–55
Jiang JX (2014) Identification and functional characterization of microRNAs involved in pollen development in Brassica campestris ssp. Chinensis. Master Dissertation. Zhejiang University, Hangzhou, China (in Chinese)
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:68–73
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Lei KJ, Liu H (2016) Research advances in plant regulatory hub miR156 and targeted. Chem Life 36:13–20 (in Chinese)
Li H, Deng Y, Wu T, Subramanian S, Yu O (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol 153:1759–1770
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Rogers K, Chen X (2013) Biogenesis, turnover and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 36:1101–1108
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specifc effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shen J (2015) The cloning and drought response of miR858 and the mechanism for it negatively regulates anthocyanin biosynthesis in tomato. Master Dissertation. Shanxi Agricultural University, Taigu, China (in Chinese)
Spanudakis E, Jackson S (2014) The role of microRNAs in the control of flowering time. J Exp Bot 65:365–380
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Wang JG, Zhang ZS (2005) Herbaceous peonies of China. China Forestry Publishing House, Beijing in Chinese)
Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X (2010a) miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 62:761–773
Wang L, Mai YX, Zhang YC, Qian L, Yang HQ (2010b) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3:794–806
Wang L, Zeng HQ, Song J, Feng SJ, Yang ZM (2015) miRNA778 and SUVH6 are involved in phosphate homeostasis in Arabidopsis. Plant Sci 238:273–285
Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X (2010) Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genom 11:246
Yang F, Cai J, Yang Y, Liu Z (2013) Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis. Plant Cell Tiss Org Cult 2:159–167
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8:R96
Zhang BH, Pan XP, Anderson TA (2006) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhao DQ, Jiang Y, Ning CL, Meng JS, Lin SS, Ding W, Tao J (2014) Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactiflora Pall.). BMC Genom 15:689
Zhao DQ, Wei MR, Liu D, Tao J (2016) Anatomical and biochemical analysis reveal the role of anthocyanins in flower coloration of herbaceous peony. Plant Physiol Biochem 102:97–106
Zhao DQ, Wei MR, Shi M, Hao ZJ, Tao J (2017) Identification and comparative profiling of miRNAs in herbaceous peony (Paeonia lactiflora Pall.) with red/yellow bicoloured flowers. Sci Rep 7:44926
Acknowledgements
This work was supported by the Natural Science Foundation of China (31,400,592), the program of key members of Yangzhou University outstanding young teachers and the Priority Academic Program Development from Jiangsu Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author declares that there is no competing interest towards the publication of this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, D., Xia, X., Wei, M. et al. Overexpression of herbaceous peony miR156e-3p improves anthocyanin accumulation in transgenic Arabidopsis thaliana lateral branches. 3 Biotech 7, 379 (2017). https://doi.org/10.1007/s13205-017-1011-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-017-1011-3