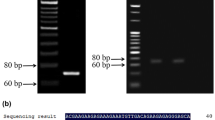
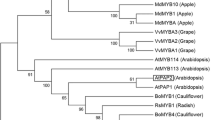
Abstract
Plant microRNAs (miRNAs) are RNAs of 20–22 nucleotides in length with sequence complementarities to specific mRNAs that are targeted for either cleavage or translational repression. They play critical regulatory roles in plant growth and development. In this study, the functionality of miR828 in the regulation of anthocyanin biosynthesis has been investigated. RNA blotting analysis has confirmed the presence of a conserved 22 nucleotide miR828 in both dicot and monocot plants. Moreover, it has revealed that miR828 is constitutively expressed in different tissues of Arabidopsis. Subsequently, a 35S:pre-miR828 construct has been created and transformed into Arabidopsis. Expression analysis has shown that levels of pre-miR828 and mature miR828 transcripts are increased, while levels of MYB75, MYB90, and MYB113 transcripts, encoding MYB transcription factors that positively regulate anthocyanin biosynthesis, are repressed in transgenic plants. As a result, anthocyanin levels are reduced, as are transcription levels of genes that are directly involved in anthocyanin biosynthesis, including PAL, CHS, CHI, F3H, F3′H, DFR, and LDOX. In addition, overexpression of miR828 in Arabidopsis inhibits transcription of yet another MYB factor, MYB82, indicating that MYB82 is likely to be involved in the anthocyanin biosynthesis pathway.





Similar content being viewed by others
Abbreviations
- PAL:
-
Phenylalanine ammonialyase
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- F3H:
-
Flavanone 3-hydroxylase
- F3′H:
-
Flavonoid 3′-hydroxylase
- DFR:
-
Dihydroflavonol reductase
- LDOX:
-
Leucoanthocyandin dioxygenase
References
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Axtell MJ, Bartel DP (2005) Antiquity of microRNAs and their target in land plant. Plant cell 17:1658–1673
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Aarabidopsis. Curr Opin Plant Biol 8:272–279
Carey CC, Strahle JT, Selinger DA, Chandler V (2004) Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotype relative to the functionally similar Transparent Testa Glabra 1 gene in Arabidopsis thaliana. Plant Cell 16:450–464
Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Gaspero DG (2007) Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine seasonal water deficit. Plant, Cell Environ 30:1381–1399
Christie PJ, Alfenito MR, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194:541–549
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7:512–520
Gao JJ, Shen XF, Zhang Z, Peng RH, Xiong AS et al (2011) The myb transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis. Plant Cell Tiss Org 106(2):235–242
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17:1376–1386
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
He J, Silva AMS, Mateus N, Freitas V (2011) Oxidative formation and structural characterisation of new α-pyranone (lactone) compounds of non-oxonium nature originated from fruit anthocyanins. Food Chem 127:984–992
Honda C, Kotoda N, Wada M, Satoru K, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Bioch 40:955–962
Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151:2120–2132
Ishide T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kate T, Tabata S, Okada K, Wada T (2007) Arabidopsis TRANSPARENT TESTA GLABRA 2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA 2 transcription in epidermal differentiation. Plant cell 19:2531–2543
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jones-Rhoades MW, Bartel DP, Bartel B (2006) Micro-RNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Li WF, Zhang SG, Han SY, Wu T, Zhang JH, Qi LW (2013) Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.). Carr Plant Cell Tiss Org 113(1):131–136
Liang G, Yang F, Yu D (2010) MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J 62:1046–1057
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605–1619
Lorenc-Kukula K, Jafra S, Oszmianski J, Szopa J (2005) Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J Agric Food Chem 53:272–281
Luo Q, Mittal A, Jia F, Rock CD (2012) An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol Biol 80:117–129
Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47:457–470
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Pelletier MK, Murrell JR, Winkel-Shirley B (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Plant Physiol 113:1437–1445
Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poething RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Gene Dev 18:2368–2379
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev 20:3407–3425
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Sarma AD, Sharma R (1999) Anthocyanin-DNA copigmentation complex: mutual protection against oxidative damage. Phytochemistry 52:1313–1318
Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140:637–646
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50:660–677
Sun C, Zhao Q, Liu DD, You CX, Hao YJ (2013) Ectopic expression of the apple Md-miRNA156 h gene regulates flower and fruit development in Arabidopsis. Plant Cell Tiss Org 112(3):343–351
Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Gene Dev 17:49–63
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16:69–79
Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166 g and its AtHD-ZIP target genes. Development 132:3657–3668
Winkel-Shirley B, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Xia R, Zhu H, An YQ, Beers E, Liu Z (2012) Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol 13(6):R47
Yang F, Liang G, Liu D, Yu D (2009) Arabidopsis miR396 mediates the development of leaves and flowers in transgenic tobacco. J Plant Biol 52(5):475–481
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30971557 to Y.Y), and Chinese Universities Scientific Fund (2011SCU11107).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, F., Cai, J., Yang, Y. et al. Overexpression of microRNA828 reduces anthocyanin accumulation in Arabidopsis . Plant Cell Tiss Organ Cult 115, 159–167 (2013). https://doi.org/10.1007/s11240-013-0349-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-013-0349-4