Abstract
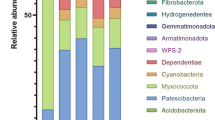
The resident microbial population responsible for lignocellulosic biomass assimilation in the gastrointestinal tract of animals is a rich source for discovering novel biocatalysts finding application in the production of value-added commodities. Herbivorous animals, such as elephants, consume a variety of lignocellulosic materials in bulk amounts to support their high energy requirements. Since the interdependence of host diet and its microbiome is well established, it is necessary to explore the potential resident microbes of obligate herbivores like elephants belonging to different age classes and habitats for mining enzymes involved in complex biomass deconstruction. In the present study, metagenomic analysis of an adult elephant fecal sample using whole-genome shotgun library preparation indicated the dominant representation of microbes belonging to the phylum Proteobacteria. Subsystem- and KEGG-based analyses revealed a high potential for carbohydrate metabolism and membrane transport. CAZy database analysis identified ~55,000 ORFs that had either catalytic domains or carbohydrate-binding modules (CBMs) in the metagenomic data set. Moreover, CBMs and carbohydrate-active enzymes (CAZymes), such as glycoside hydrolases (GHs), glycosyltransferases (GTs), carbohydrate esterases (CEs) were most abundant in microbes of phylum Proteobacteria, and among them, the majority of GHs and GTs were from Bacillus subtilis and Escherichia coli. A comparative GH analysis with other gut metagenomic datasets of herbivorous animals revealed the presence of several unique GHs of the β-glucosidase, endoglucanase, and exoglucanase families thus providing a comprehensive understanding of the diverse CAZymes present in the gut microbiome of an adult elephant.





Similar content being viewed by others
References
Biver S, Vandenbol M (2013) Characterization of three new carboxylic ester hydrolases isolated by functional screening of a forest soil metagenomic library. J Ind Microbiol Biotechnol 40:191–200. https://doi.org/10.1007/s10295-012-1217-7
Brulc JM et al (2009) Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A 106:1948–1953. https://doi.org/10.1073/pnas.0806191105
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. https://doi.org/10.1093/nar/gkn663
Chen B, Teh BS, Sun C, Hu S, Lu X, Boland W, Shao Y (2016) Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci Rep 6:29505. https://doi.org/10.1038/srep29505
Emami K, Topakas E, Nagy T, Henshaw J, Jackson KA, Nelson KE, Mongodin EF, Murray JW, Lewis RJ, Gilbert HJ (2009) Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J Biol Chem 284:1086–1096. https://doi.org/10.1074/jbc.M805100200
Ezer A et al (2008) Cell surface enzyme attachment is mediated by family 37 carbohydrate-binding modules, unique to Ruminococcus albus. J Bacteriol 190:8220–8222. https://doi.org/10.1128/JB.00609-08
Fatima F, Pathak N, Rastogi Verma S (2014) An improved method for soil DNA extraction to study the microbial assortment within rhizospheric region. Mol Biol Int 2014:518960. https://doi.org/10.1155/2014/518960
Geddes CC, Nieves IU, Ingram LO (2011) Advances in ethanol production. Curr Opin Biotechnol 22:312–319. https://doi.org/10.1016/j.copbio.2011.04.012
Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T (2010) Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. https://doi.org/10.1111/j.1365-2958.2009.06793.x
Gupta VK et al (2016) Fungal enzymes for bio-products from sustainable and waste biomass. Trends Biochem Sci 41:633–645. https://doi.org/10.1016/j.tibs.2016.04.006
Gysler C, Harmsen JAM, Kester HCM, Visser J, Heim J (1990) Isolation and structure of the pectin lyase D-encoding gene from Aspergillus niger. Gene 89:101–108. https://doi.org/10.1016/0378-1119(90)90211-9
Hayashi K et al (1997) Pectinolytic enzymes from Pseudomonas marginalis maff 03-01173. Phytochemistry 45:1359–1363. https://doi.org/10.1016/s0031-9422(97)00191-x
Heredia A, Jimenez A, Guillen R (1995) Composition of plant cell walls. Zeitschrift fur Lebensmittel-Untersuchung und -Forschung 200:24–31
Hess M et al (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. https://doi.org/10.1126/science.1200387
Ichinose H et al (2005) An exo-beta-1,3-galactanase having a novel beta-1,3-galactan-binding module from Phanerochaete chrysosporium. J Biol Chem 280:25820–25829. https://doi.org/10.1074/jbc.M501024200
Ilmberger N, Güllert S, Dannenberg J, Rabausch U, Torres J, Wemheuer B, Alawi M, Poehlein A, Chow J, Turaev D, Rattei T, Schmeisser C, Salomon J, Olsen PB, Daniel R, Grundhoff A, Borchert MS, Streit WR (2014) A comparative metagenome survey of the fecal microbiota of a breast- and a plant-fed Asian elephant reveals an unexpectedly high diversity of glycoside hydrolase family enzymes. PLoS One 9:e106707. https://doi.org/10.1371/journal.pone.0106707
Jin Y et al (2016) A beta-Mannanase with a lysozyme-like fold and a novel molecular catalytic mechanism. ACS Cent Sci 2:896–903. https://doi.org/10.1021/acscentsci.6b00232
Katayama Y, Nishikawa S, Murayama A, Yamasaki M, Morohoshi N, Haraguchi T (1988) The metabolism of biphenyl structures in lignin by the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett 233:129–133. https://doi.org/10.1016/0014-5793(88)81369-3
Lafond M, Navarro D, Haon M, Couturier M, Berrin JG (2012) Characterization of a broad-specificity beta-glucanase acting on beta-(1,3)-, beta-(1,4)-, and beta-(1,6)-glucans that defines a new glycoside hydrolase family. Appl Environ Microbiol 78:8540–8546. https://doi.org/10.1128/AEM.02572-12
Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. https://doi.org/10.1186/1754-6834-6-41
Liu QP et al (2007) Bacterial glycosidases for the production of universal red blood cells. Nat Biotechnol 25:454–464. https://doi.org/10.1038/nbt1298
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma 9:386. https://doi.org/10.1186/1471-2105-9-386
Mielenz JR (2001) Ethanol production from biomass: technology and commercialization status. Curr Opin Microbiol 4:324–329
Montella S, Ventorino V, Lombard V, Henrissat B, Pepe O, Faraco V (2017) Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci Rep 7:42623. https://doi.org/10.1038/srep42623
Moreira LR, Filho EX (2016) Insights into the mechanism of enzymatic hydrolysis of xylan Applied microbiology and biotechnology 100:5205–5214. https://doi.org/10.1007/s00253-016-7555-z
Nagaraja TG (2016) Microbiology of the rumen. In: Millen D, De Beni Arrigoni M, Lauritano Pacheco R (eds) Rumenology. Springer Cham pp. 39–61. https://doi.org/10.1007/978-3-319-30533-2_2
Natarajan VP, Zhang X, Morono Y, Inagaki F, Wang F (2016) A modified SDS-based DNA extraction method for high quality environmental DNA from seafloor environments. Front Microbiol 7:986. https://doi.org/10.3389/fmicb.2016.00986
Peng B, Huang S, Liu T, Geng A (2015) Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb Cell Fact 14:70. https://doi.org/10.1186/s12934-015-0253-1
Pope PB et al (2010) Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci U S A 107:14793–14798. https://doi.org/10.1073/pnas.1005297107
Prade RA, Zhan D, Ayoubi P, Mort AJ (1999) Pectins, pectinases and plant-microbe interactions. Biotechnol Genet Eng Rev 16:361–392. https://doi.org/10.1080/02648725.1999.10647984
Scully ED, Geib SM, Hoover K, Tien M, Tringe SG, Barry KW, Glavina del Rio T, Chovatia M, Herr JR, Carlson JE (2013) Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS One 8:e73827. https://doi.org/10.1371/journal.pone.0073827
Sharma A, Tewari R, Rana SS, Soni R, Soni SK (2016) Cellulases: classification, methods of determination and industrial applications. Appl Biochem Biotechnol 179:1346–1380. https://doi.org/10.1007/s12010-016-2070-3
Suen G, Teiling C, Li L, Holt C, Abouheif E, Bornberg-Bauer E, Bouffard P, Caldera EJ, Cash E, Cavanaugh A, Denas O, Elhaik E, Favé MJ, Gadau J, Gibson JD, Graur D, Grubbs KJ, Hagen DE, Harkins TT, Helmkampf M, Hu H, Johnson BR, Kim J, Marsh SE, Moeller JA, Muñoz-Torres MC, Murphy MC, Naughton MC, Nigam S, Overson R, Rajakumar R, Reese JT, Scott JJ, Smith CR, Tao S, Tsutsui ND, Viljakainen L, Wissler L, Yandell MD, Zimmer F, Taylor J, Slater SC, Clifton SW, Warren WC, Elsik CG, Smith CD, Weinstock GM, Gerardo NM, Currie CR (2011) The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet 7:e1002007. https://doi.org/10.1371/journal.pgen.1002007
Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ (2006) Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol 8:1997–2011. https://doi.org/10.1111/j.1462-2920.2006.01080.x
Van Hoven AW, Prins RA, Lankhorst (1981) Fermentative digestion in the African elephant. Afr J Wildl Res 11:78–88
Villares A, Moreau C, Bennati-Granier C, Garajova S, Foucat L, Falourd X, Saake B, Berrin JG, Cathala B (2017) Lytic polysaccharide monooxygenases disrupt the cellulose fibers structure. Sci Rep 7:40262. https://doi.org/10.1038/srep40262
Walterson AM, Stavrinides J (2015) Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev 39:968–984. https://doi.org/10.1093/femsre/fuv027
Warnecke F et al (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. https://doi.org/10.1038/nature06269
Wohlkonig A, Huet J, Looze Y, Wintjens R (2010) Structural relationships in the lysozyme superfamily: significant evidence for glycoside hydrolase signature motifs. PLoS One 5:e15388. https://doi.org/10.1371/journal.pone.0015388
Xu B, Xu W, Li J, Dai L, Xiong C, Tang X, Yang Y, Mu Y, Zhou J, Ding J, Wu Q, Huang Z (2015) Metagenomic analysis of the Rhinopithecus bieti fecal microbiome reveals a broad diversity of bacterial and glycoside hydrolase profiles related to lignocellulose degradation. BMC Genom 16:174. https://doi.org/10.1186/s12864-015-1378-7
Yun JH et al (2014) Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol 80:5254–5264. https://doi.org/10.1128/AEM.01226-14
Zhu L, Wu Q, Dai J, Zhang S, Wei F (2011) Evidence of cellulose metabolism by the giant panda gut microbiome. Proc Natl Acad Sci U S A 108:17714–17719. https://doi.org/10.1073/pnas.1017956108
Acknowledgements
We are thankful to National Zoological Park, New Delhi, India, for providing elephant fecal samples. NAG acknowledges ICGEB, New Delhi and Department of Biotechnology (DBT), Government of India for financial support (Grant No.: BT/PB/Centre/03/ICGEB/2011-PhaseII).
Funding
This work was supported in part by DST Ramanujan Fellowship grant (SR/S2/RJN-07/2012) and DBT grant (BT/PR10684/PBD/26/403/2013).
Author information
Authors and Affiliations
Contributions
NAG and JS1 contributed to the initial design of the research. JS1 and SJ conducted the experiments, and MV and JS1 performed the bioinformatics analyses with guidance from DG, MK and NAG. The initial draft of the manuscript was prepared by NAG, JS1, FM and MV, scientific assistance and correction was done by JS2 and all the authors contributed to the subsequent stages of manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Reviewer’s access
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1.30 mb)
Rights and permissions
About this article
Cite this article
Jakeer, S., Varma, M., Sharma, J. et al. Metagenomic analysis of the fecal microbiome of an adult elephant reveals the diversity of CAZymes related to lignocellulosic biomass degradation. Symbiosis 81, 209–222 (2020). https://doi.org/10.1007/s13199-020-00695-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-020-00695-8