Abstract
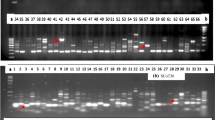
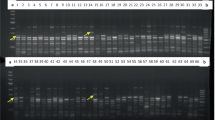
Assessment of genetic diversity of Bambara groundnut (Vigna subterranea (L.) Verdcourt) accessions from Nigeria using informative molecular markers has become imperative for their genetic improvement and conservation. Comparative analysis of 30 V. subterranea from different locations in Nigeria was investigated using Directed Amplified Minisatellite DNA (DAMD) and Start codon targeted (SCoT) markers. The DNA was extracted using CTAB method for amplification. Both markers resolved the accessions into eight major groups using dendrograms and six clusters by principal component analysis. Alleles of 25 and 53 were obtained with DAMD and SCoT, respectively. Mean alleles, gene diversity, and polymorphic information content detected with DAMD were 10.2000, 0.6950, and 0.6600, while SCoT yielded 16.200, 0.847, and 0.836, respectively. Polymorphic loci were 130 and 142 in DAMD and SCoT, respectively. Both markers produced high polymorphism of 60.00-80% and 40.33-96.67% in DAMD and SCoT, respectively. Effective alleles (Ne) in both markers (DAMD: 1.1828-1.5927; SCoT: 1.1830-1.6779) were high. The Nei’s value (H) ranged from 0.1959-0.3238 in DAMD and 0.1533-0.3782 in SCoT. The Shannon’s information index (I) obtained from DAMD and SCoT were 0.3281-0.4635 and 0.1420-0.5557, respectively. Total gene diversity (Ht), diversity within population (Hs), coefficient of gene differentiation (Gst), and estimate of gene flow (Nm) from DAMD were 0.3304, 0.2673, 0.1907, and 2.1215, while SCoT had 0.3927, 0.3163, 0.1945, and 2.0713, respectively. Our study demonstrated that SCoT markers are more competent than DAMD and should be integrated in the exploration of genetic diversity and selection of unique accessions for improvement and utilization of V. subterranea.
Similar content being viewed by others
References
Abarshi MM, Mohammed IU, Wasswa P, Hillocks RJ, Holta J, Legg JP, Seal SE, Maruthi MN. 2010. Optimization of diagnostic RT-PCR protocols and sampling procedures for the reliable and cost-effective detection of Cassava brown streak virus. J. Virol. Methods 163: 353–359
Agarwal M, Shrivastava N, Padh H. 2008. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 27: 617–631
Amadou HI, Bebeli PJ, Kaltsikes PJ. 2001. Genetic diversity in Bambara groundnut (Vigna subterranea L.) germplasm revealed by RAPD markers. Genome 44: 995–999
Amarteifio JO, Tibe O, Njogu RM. 2006. The mineral composition of Bambara groundnut (Vigna subterranea [L.] Verdc.) grown in Southern Africa. Afri. J. Biotechnol. 5: 2408–2411
Amirmoradi B, Talebi R, Karami E. 2012. Comparison of genetic diversity and differentiation among annual Cicer species using Start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Sys. Evol. Springer 12: 669–696
Appropedia. 2016. Food crops. http://www.appropedia.org/Food_crops.
Aremu MO, Olaofe O, Akintayo ET. 2006. Chemical Composition and Physicochemical Characteristics of two varieties of Bambara groundnut (Vigna subterrenea). J. Appl. Sci. 6: 1900–1903
Bacchetta G, Fenu G, Mattana E. 2011. Genetic variability of the narrow endemic Rhamnus persicifolia Moris (Rhamnaceae) and its implications for conservation. Biochem. Sys. Ecol. 39: 477–484
Bamishaiye OM, Adegbola JA, Bamishaiye EI. 2011. Bambara groundnut: an under-utilized nut in Africa. Adv. Agri. Biotechnol. 1: 60–72
Basu S, Mayes S, Davey M, Roberts JA, Azam-Ali SN, Mithen R, Pasquet RS. 2007. Inheritance of domestication’traits in Bambara groundnut (Vigna subterranea [L.] Verdc.). Euphytica 157(1–2): 59–68
Berchie J, Adu-Dapaah H, Dankyi AA, Plahar WA, Nelson-Quartey F, Haleegoah J, Asafu-Agyei JN, Addo JK. 2010. Practices and constraints in Bambara groundnuts production, marketing and consumption in the brong ahafo and upper-East Regions of Ghana. J. Agron. 9(3): 111–118
Bilgen M, Karaca M, Onus AN, Ince AG. 2004. A software program combining sequence motif searches with keywords for finding repeats containing DNA sequences. Bioinformatics 20: 3379–3386
Brink M, Ramolemana GM, Sibuga KP. 2006. Vigna subterranea (L.) Verdc. Record from Protabase. Brink, M. and Belay, G. (Editors). Plant Resources of Tropical Africa / Ressources végétales de l’Afrique tropicale. (PROTA) Wageningen, Netherlands, p 27
Collard BCY, Mackill DJ. 2009. Start Codon Targeted (SCoT) polymorphism: a simple novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Reports 27: 86–93
Dansi A, Vodouhe R, Azokpota P. 2012. Diversity of the neglected and underutilized crop species of importance in Benin. Sci. World J. pp 932–947
DPP. 2011. Directorate Plant Production of guidelines for Bambara groundnuts. Pretoria, South Africa: Department of Agriculture, Forestry and Fisheries pp 1–10
Eitzinger J, Orlandini S, Stefanski R, Naylor REL. 2010. Climate change and agriculture: Introductory Editorial. J. Agri. Sci. 148: 499–500
Eltayeb AR, Ali SM, Abou-Arab AO, Abu-Salem AA. 2011. Chemical composition and functional properties of flour and protein isolate extracted from Bambara groundnut (Vigna subterranea). Afri. J. Food Sci. 5: 82–90
FAO. 2016. Food and Agriculture Organization of the United States: Traditional Crop of the Month: Bambara groundnut. http://www.fao.org/traditional-crops/bambaragroundnut/en/.
Fatehi R, Talebi R, Fayyaz F. 2011. Characterizations of Iranian landrace wheat accessions by inter-simple sequence repeat (ISSR) markers. J. Appl. Environ. Biol. Sci. 1(10): 423–436
Gorji AM, Poczai P, Polgar Z, Taller J. 2011. Efficiency of arbitrarily amplified dominant markers (SCOT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am. J. Potato Res. 88: 226–237
Gupta PK, Rustgi S. 2004. Molecular markers from the transcribed/expressed region of the genome in higher plants. Funct. Integr. Genom. 4: 139–162
Gwag J, Dixit A, Park YJ, Ma KH, Kwon SJ, Cho GT, Lee GA, Lee SY, Kang HK, Lee SH. 2010. Assessment of genetic diversity and population structure in mungbean. Genes Genom. 32: 299–308
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R. 2015. Characterization of genetic diversity in chickpea using SSR markers, Start Codon Targeted Polymorphism (SCoT) and Conserved DNA-Derived Polymorphism (CDDP). Physiol. Mol. Biol. Plants 21(3): 365–373
Hamidi H, Talebi R, Keshavarz F. 2014. Comparative efficiency of functional gene-based markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived Polymorphism (CDDP) with ISSR markers for diagnostic fingerprinting in wheat (Triticum aestivum L.). Cereal Res. Commun. doi: 10.1556/CRC.2014.0010
Hamrick JL. 1989. Isozymes and the analysis of genetic structure in plant population. In: DE Soltis, PS Soltis, Eds., Isozymes in Plant Biology. Dioscorides Press, Portland, pp 87–105
Heath DD, Iwama GK, Devlin RH. 1993. PCR primed with VNTR core sequences yields species specific patterns and hypervariable probes. Nucleic Acids Res. 21: 5782–5785
Ho WK, Chai HH, Kendabie P, Ahmad NS, Jani J, Massawe F, Kilian A, Mayes S. 2017. Integrating genetic maps in Bambara groundnut [Vigna subterranea (L.) Verdc.] and their syntenic relationships among closely related legumes. BMC 18: 192
IITA. 2017. International Institute of Tropical Agriculture. (http://my.iita.org/accession2/collection.jspx;jsessionid=58CA6CCC459D93C7504F4B2076463FF8?id=8, IITA
Jeffreys AJ, Wilson V, Thein SL. 1985. Hypervariable ‘minisatellite’ regions in human DNA. Nature 314: 67–73
Karron JD. 1991. Patterns of genetic variation and breeding systems in rare plant species. In: D Falk, K Holsinger, Eds., Genetics and Conservation of Rare Plants, New York, Oxford University Press, pp 87–98
Khan MA, Ammar MH, Migdadi HM, El-Harty EH, Alfaifi SA, Farooq M, Alghamdi SS. 2016. Field performance and genetic diversity of chickpea genotypes. Inter. J. Agri. Biol. 18: 683–688
Lankau RA, Strauss SY. 2008. Community complexity drives patterns of natural selection on a chemical defense of Brassica nigra. Am. Nat. 171: 150–161
Li K, Chen L, Feng Y. 2014. High genetic diversity but limited gene flow among remnant and fragmented natural populations of Liriodendron chinense Sarg. Biochem. Sys. Ecol. 54: 230–236
Mooney KA, Agrawal AA. 2008. Plant genotype shapes ant–aphid interactions: Implications for community structure and indirect plant defense. Am. Nat. 171: 195–205
Nadembega S. 2016. Food production and food security in Burkina Faso: Case of the voandzou in three communes of the three agro-ecological zones. Diploma of Master 2, Université Catholique de l’Afrique de l’Ouest, Bobo Dioulasso, p 90
Ndidi US, Ndidi CU, Olagunju A, Muhammad A, Billy FG, Okpe O. 2014. Proximate, antinutrients and mineral composition of raw and processed (boiled and roasted) Sphenostylis stenocarpa seeds from Southern Kaduna, Northwest Nigeria. ISRN 10: 1155–2808
Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590
Ntundu WH, Bach IC, Christiansen JL, Andersen SB. 2003. Analysis of genetic diversity in bambara groundnut [Vigna subterranea (L.) Verdc] landraces using amplified fragment length polymorphism (AFLP) markers. Afri. J. Biotechnol. 3(4): 220–225
Odongo FO, Oyoo ME, Wasike V, Owuche JO, Karanja L, Korir P. 2015. Genetic diversity of Bambara groundnut (Vigna subterranea (L.) Verdc.) landraces in Kenya using microsatellite markers. Afri. J. Biotechnol. 14(4): 283–291
Okonkwo SL, Opara MF. 2010. The analysis of bambara nut (Voandzeia Subterranea (L.) Thouars) for sustainability in Africa. Res. J. Appl. Sci. 5: 394–396
Okpuzor J, Ogbunugafor H, Okafor U, Sofidiya M. 2010. Identification of protein types in Bambara nut seeds: perspectives for dietary protein supply in developing countries. EXCLI J. 9: 17–28
Olukolu BA, Mayes S, Stadler F, Fawole I, Dominique D, Azam ASN, Abott GA, Kole C. 2012. Genetic diversity in Bambara groundnut (Vigna subterranea (L.) Verdc.) as revealed by phenotypic descriptors and DArT marker analysis. Genet. Resour. Crop Evol. 59: 347–358
Ojuederie BO, Igwe DO, Okuofu SI, Faloye B. 2013. Assessment of Genetic Diversity in some Moringa oleifera Lam. Landraces from Western Nigeria using RAPD markers. Afri. J. Plant Sci. Biotechnol. 7(1): 15–20
Ouoba A, Ouedraogo M, Sawadogo M, Nadembega S. 2016. Aperçu de la culture du voandzou (Vigna subterranea (L.) Verdcourt) au Burkina Faso: enjeux et perspectives d’amélioration de sa productivité. Inter. J. Biol. Chem. Sci. 10(2): 652–665
Pakseresht F, Talebi R, Karami E. 2013. Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of chickpea (Cicer arietinum L.) landraces genotypes collected from north-west of Iran. Physiol. Mol. Biol. Plants 19(4): 563–574
Poczai P. 2011. Molecular genetic studies on complex evolutionary processes in Archae solanum (Solanum, Solanaceae), PhD thesis, University of Pannonia, pp 126
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JPT, Hyvönen J. 2013. Advances in plant gene-targeted and functional markers: a review. Plant Meth. 9: 6
Rao VR, Hodgkin T. 2002. Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tiss. Org. Cult. 68: 1–19
Roussel V, Koenig J, Beckert M, Balfourier F. 2004. Molecular diversity in French bread wheat accessions related to temporal trends and breeding programmes. Theor. Appl. Genet. 108: 920–930
Rungnoi O, Suwanprasert J, Somta P, Srinives P. 2012. Molecular genetic diversity of Bambara groundnut (Vigna subterranea (L.) Verdc.) revealed by RAPD and ISSR marker analysis. SABRAO. J. Breed. Genet. 44(1): 87–101
Saeed A, Hovsepyan H, Darvishzadeh R, Imtiaz M, Panguluri SK, Nazaryan R. 2011. Genetic diversity of Iranian accessions, improved lines of chickpea (Cicer arietinum L.) and their wild relatives by using simple sequence repeats. Plant Mol. Biol. Rep. 29: 848–858
Singh A, Dikshit HK, Jain N, Singh D, Yadav RN. 2014. Efficiency of SSR, ISSR and RAPD markers in molecular characterization of mungbean and other Vigna species. Indiana J. Biotechnol. 13: 81–88
Slatkin M, Barton NH. 1989. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368
Somers DJ, Demmon G. 2002. Identification of repetitive, genome-specific probes in crucifer oilseed species. Genome 45: 485–492
Somta P, Chankaew S, Rungnoi O, Srinives P. 2017. Genetic diversity of the Bambara groundnut (Vigna subterranea (L.) Verdc.) as assessed by SSR markers. Genome 54: 898–910
Souframanien J, Gopalakrishna T. 2004. A comparative analysis of genetic diversity in black gram genotypes using RAPD and ISSR markers. Theor. Appl. Genet. 109: 1687–1693
Stephens JM. 2013. Bambara Groundnut Voandzeia subterranea (L.) Thouars. Gainesville, Florida: University of Florida; 2012. http://edis.ifas.ufl.edu/mv014
Suthar KP, Bhatnagar R, Shukla YM, Suthar VP, Kadam SD, Patel NJ. 2012. Genetic diversity assessment in Chickpea genotypes using STMS. Legume Res. 35(4): 285–293
Talebi R, Fayaz R, Mardi M, Pirsyedi SM, Naji AM. 2008. Genetic relationships among chickpea (Cicer arietinum) elite lines based on RAPD and agronomic markers. Inter. J. Agri. Biol. 8: 301–305
Twyford AD, Kidner CK, Harrison N, Ennos RA. 2013. Population history and seed dispersal in widespread Central American Begonia species (Begoniaceae) inferred from plastome-derived microsatellite markers. Bot. J. Linn. Soc. 171: 260–276
Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda CLL. 2008. Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 8: 106. doi: 10.1186/1471-2229-8-106
Vurayai R, Emongor V, Moseki B. 2011. Effect of water stress imposed at different growth and developmental stages on morphological traits and yield of bambara groundnuts (Vigna subterranea L. Verdc). Am. J. Plant Physiol. 6: 17–27
Wright S. 1951. The genetic structure of populations. Ann. Eugenics 15: 313–354
Yeh FC, Yang RC, Boyle T. 1999. Popgene Version 1.31. Microsoft Window-based Freeware for Population Genetic Analysis. Department of Renewable Resources, University of Alberta, Edmonton, AB Canada
Yifru T, Hammer K, Huang XQ, Roder MS. 2006. Regional patterns of microsatellite diversity in Ethiopian tetraploid wheat accessions. Plant Breed. 125: 125–130
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Igwe, D.O., Afiukwa, C.A. Competency assessment of directed amplified minisatellite DNA and start codon targeted markers for genetic diversity study in accessions of Vigna subterranea (L.) Verdcourt. J. Crop Sci. Biotechnol. 20, 263–278 (2017). https://doi.org/10.1007/s12892-017-0119-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-017-0119-0