Abstract
The aim of our study was to prepare multifunctional, biocompatible nanoparticles for site-specific drug delivery. Hydrophilic nanoparticles with surface-adorned amine, carboxyl, or aldehyde groups, to be later used for bio-conjugation, were designed using phosphonomethyl iminodiacetic acid (PMIDA) as the coupling agent. These PMIDA-coated cobalt oxide nanoparticles (PMIDA-CoO) were further functionalized with folic acid (FA), using simple technique. The particles show excellent aqueous dispersion stability in physiological pH without any deterioration in hydrodynamic size. The cytotoxicity and internalization efficiency of these nanocarriers have been evaluated on folate receptor over expressed KB and folate receptor lower expressed KG1a cells. Anticancer drugs such as doxorubicin and methotrexate were successfully attached to the folic acid-decoded PMIDA-CoO nanoparticles by simple reactions. Anticancer drug-loaded nanoparticles (FA-PMIDA-CoO) exhibit elevated cytotoxicity and induce apoptosis in cancer cells, which were confirmed by flow cytometry. Fluorescence microscopy study shows the higher amount of internalization of the noncomplex by KB cells, which clearly demonstrated that cells overexpressing the human folate receptor internalized a higher level of these nanoparticles–folate conjugates than folate receptor-negative control cells.
Similar content being viewed by others
1 Introduction
The uses of nanoparticles (NPs) with functional properties have been widely used in a broad range of bio-applications, like drug and gene delivery, cell and tissue engineering, diagnostic and therapeutic purposes, etc. (Panyam and Labhasetwar 2003; Marin et al. 2005). Among these applications, the field of drug delivery by NPs with specific and rapid internalization into a target cell has immense promise (Maeda et al. 2009; Faraji and Wipf 2009). Folic acid is a member of vitamin B family and plays an essential role in cell survival by participating in the biosynthesis of nucleic acids (Antony 1996). This essential vitamin is also a high-affinity ligand that enhances the differential specificity of conjugated anticancer drugs by targeting folate receptor (FR)-positive cancer cells (Leamon and Reddy 2004). The folic acid (FA)-conjugated drugs can actively internalize bound folates and folate-conjugated compounds via receptor-mediated endocytosis (Kamen and Capdevila 1986; Leamon and Low 1991). It has been found that FR is upregulated in more than 90 % of ovarian carcinomas. It is also found at high to moderate levels in the kidney, brain, lung, and breast carcinomas, while it occurs at very low levels in most normal tissues (Kamen and Smith 2004). The FR density also appears to increase as the stage of the cancer increases (Elnakat and Ratnam 2004). Various types of anticancer drugs were conjugated and evaluated for their biological activity (Low and Antony 2004).
Metal NPs are the most interesting chemical elements used as NPs because of their potential in a wide range of biotechnological applications, especially in advanced biomedical application (Liong et al. 2008). Cobalt (Co) which is mainly used as cobalt oxide has an organometal compound or a biopolymer (Wang et al. 2005). In spite of its physiological role as a cofactor of vitamin B12, cobalt cannot be regarded only as an essential element. Cobalt oxide NPs have anticancer activity (Papis et al. 2009) through surface modification (Chattopadhyay et al. 2012). Therefore, in the present paper, we have focused our interest on the study of receptor-targeted delivery of modified cobalt oxide (CoO) NPs. We used one folate receptor over expressing cancer cell line KB (Lee and Low 1994) and one folate receptor lower expressing cancer cell line KG-1A (Pan et al. 2002). This present study was aimed to develop novel and inexpensive NPs which have anticancer activity to cancer cell in vitro as well as serve as a drug carrier.
2 Materials and methods
2.1 Chemicals and reagents
Phosphonomethyl iminodiacetic acid (PMIDA), cobalt chloride (CoCl2·3H2O), N-hydroxysuccinimide (NHS), 2,2-(ethylenedioxy)-bis-(ethylamine) (EDBE), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), propidium iodide (PI), RNase A, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT reagent), rhodamine B (RhB), Histopaque 1077, ethidium bromide (EtBr), folic acid, doxorubicin (DOX), and methotrexate (MTX) were procured from Sigma (St. Louis, MO, USA). Minimum essential medium (MEM), RPMI 1640, fetal bovine serum (FBS), penicillin, streptomycin, sodium chloride (NaCl), sodium carbonate, sucrose, Hank’s balanced salt solution, and ethylenediaminetetraacetate were purchased from HiMedia, India. Tris–hydrochloric acid (HCl), Tris buffer, KH2PO4, K2HPO4, HCl, formaldehyde, alcohol, and other chemicals were procured from Merck Ltd., SRL Pvt. Ltd., Mumbai, India. Commercially available dimethyl sulfoxide (DMSO) was procured from HiMedia, India and was purified by vacuum distillation over KOH. All other chemicals were from Merck Ltd., SRL Pvt., Ltd., and were of the highest purity grade available.
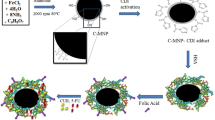
2.2 Synthesis of PMIDA-coated cobalt oxide NPs and folic acid-coated NPs
CoO NPs were synthesized by calcinated method (Chattopadhyay et al. 2012) followed by conjugation with PMIDA. To conjugate the nanoparticles with folic acid, FA was dissolved in ∼10 ml of DMSO (0.025 mM) due to the limited solubility of FA in aqueous medium. FA-EDBE was synthesized according to our previously reported protocol (Das et al. 2008). The resulting solution was then mixed with an aqueous solution of EDC (75 mM) and NHS (15 mM). The pH of the solution was then adjusted to ∼8 by dropwise addition of pyridine. An aqueous dispersion of ∼10 mg of PMIDA-functionalized nanoparticles was added to it and the reaction mixture was stirred overnight at 37 °C in dark. Following FA conjugation, the modified particles were again isolated with a rare earth magnet, washed three times with deionized water, and redispersed in PBS (Das et al. 2008).
2.3 Preparation of folic acid-coated fluorescent NPs
Five milligrams of folic acid-decorated cobalt oxide nanoparticles was dispersed in 5 ml distilled water. RhB was added with FA-PMIDA-CoO NPs. The resulting suspension was stirred in the dark for 12 h. Particles were recovered by centrifugation and washed thoroughly with Millipore water (Rho-FA-PMIDA-CoO NPs).
2.4 Preparation of DOX-loaded FA-PMIDA-CoO nanoparticles
FA-PMIDA-CoO nanoparticles immobilized with DOX were prepared through the following process. The DOX (0.5 mg/ml) in water solution was introduced in a nanoparticles suspension (1 mg/ml) and stirred for 24 h. The suspension was then centrifuged at 15,000 rpm for 20 min to precipitate the nanoparticles and decanted. The process was repeated three times to remove any free DOX in the suspension. The amount of loaded DOX was measured by spectrophotometrically at 481 nm with a UV-1800 spectrophotometer (Shimadzu) (Sahu et al. 2010). The percentage of encapsulation (EE%) was calculated from following equation:
2.5 Preparation of MTX-loaded FA-PMIDA-CoO nanoparticles
FA-PMIDA-CoO nanoparticles immobilized with MTX were prepared through the following process. The MTX (conc. 0.5 mg/ml) in water solution was introduced in a nanoparticle suspension (1 mg/ml) and stirred for 24 h. The suspension was then centrifuged at 15,000 rpm for 20 min to precipitate the nanoparticles and decanted. The process was repeated three times to remove any free MTX in the suspension. The amount of loaded MTX was measured spectrophotometrically at 270 nm with a UV-1800 spectrophotometer (Shimadzu) (Sahu et al. 2010). The percentage of encapsulation (EE%) was calculated from following equation:
2.6 In vitro drug release
In order to study the DOX and MTX release behavior, 2.5 mg of DOX-FA-PMIDA-CoO NPs and MTX-FA-PMIDA-CoO NPs (0.5 mg/ml) was suspended in 5 ml of PBS. The solution pH was adjusted to 3, 5, and 7.4 using 0.1 M HCl and NaOH. Following incubation for 8, 12, 24, and 48 h, the nanoparticle suspensions were isolated and the MTX cleaved from nanoparticles was then quantified spectrophotometrically at 481 and 270 nm, respectively (Mohapatra et al. 2011).
2.7 Characterization
2.7.1 Characterization of FA-PMIDA-CoO NP
DLS analysis
Dynamic light scattering (DLS) analysis was done by Zetasizer Nano ZS (Malvern Instruments) according to the method of Chakraborty et al. (2010) with some modifications. The NPs were (100 μg/ml) sonicated for 2 min and dynamic particle sizes were measured suspending two drops of aqueous suspension of NPs in 10 ml of Millipore water. When particle was completely dispersed in water, then particles were analyzed with a dynamic light scattering analyzer. The experiments were repeated several times to get average size of nanoparticles.
SEM micrograph
The particle size and microstructure were studied by scanning electron microscopy (Hitachi S-3400N) according to the method of Ghosh et al. (2011) with some modifications. In brief, FA-PMIDA-CoO NPs were suspended in deionized water at a concentration of 1 mg/ml then the sample was sonicated using a sonicator bath until the sample formed a homogenous suspension. For size measurement, sonicated stock solution of all cobalt oxide NPs (0.5 mg/ml) was diluted 20 times. SEM was used to characterize the size and shape of the cobalt oxide NPs. A drop of aqueous cobalt oxide NP suspension was placed onto carbon-coated copper grid and this was dried in air to get SEM image.
FTIR analysis of folic acid conjugation
The conjugation of folic acid (FA) with PMIDA-CoO NPs was investigated by Fourier transform infrared (FTIR) model PerkinElmer Spectrum RXIFT-IR System according to the method of Ghosh et al. (2011) with some modifications. In brief, 1.0 mg sticky mass of FA-PMIDA-CoO NPs with 100 μl KBr medium and a thin film was prepared on the NaCl plate by drop casting method and under atmosphere separately. The FTIR value was taken within 400 cm−1 to 4000 cm−1 (wave numbers).
Surface chemistry of drug-loaded FA-PMIDA-CoO NP
The conjugation of drug with FA-PMIDA-CoO NPs was investigated by the FTIR with a model Thermo Nicolet Nexus model 870 according to the method of Ghosh et al. (2011) with some modifications. In brief, 1.0 mg sticky mass of DOX-FA-PMIDA-CoO NPs and MTX-FA-PMIDA-CoO NPs with 100 μl KBr medium and a thin film was prepared on the NaCl plate by drop casting method and under atmosphere separately. The FTIR value was taken within 500 to 4,000 cm−1 (wave numbers).
2.8 Solubility and stability of FA-PMIDA-CoO NPs
Stability assessments were performed at different pH of phosphate buffer varying from pH 3 to 7.4. For all measurements, the concentrations of particles were maintained at 3 mg/ml and folic acid release was measured spectrophotometrically at 270 nm.
2.9 Plasma protein binding assay
Plasma protein binding was determined by using human plasma according to Patila et al. (2007). Human plasma samples were obtained from eight healthy individuals according to institutional bioethics approval. Two milligrams per milliliter of nano-FA-PMIDA-CoO was mixed with 5 ml of 50 mM PBS and 0.5 ml of human plasma (8 mg/ml) was added together and stirred with vigorously within a shaking incubator for 24 h at 37 °C. The nanoparticles were centrifuged at 10,000 rpm for 10 min and sup was used to determine the protein concentration by Lowry et al. (1951). Plasma without NPs was used as a control to ensure that there was no protein precipitation.
2.10 Separation of PBMCs and macrophages
Fasting blood samples were collected from all groups of individuals satisfying the Helsinki protocol. The lymphocytes were isolated from heparinized blood samples according to the method of Hudson and Hay (1991). Blood was diluted with phosphate-buffered saline (pH 7.0) in equal ratio and then layered very carefully on the density gradient (Histopaque 1077) in 1:2 ratio and centrifuged at 1,400 rpm for 40 min, and the white milky layer of mononuclear cells was carefully removed and cultured in RPMI 1640 medium supplemented with 10 % fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin, 4 mM l-glutamine under 5 % CO2, and 95 % humidified atmosphere at 37 °C for 2 h. The non-adherent layer of the cultured cells was washed twice with the PBS and centrifuged at 2,000 rpm for 5 min to get the required pellet of PBMCs. The adherent layer of the cultured cells was treated with 0.25 % trypsin in PBS for 3 min at 37 °C, washed twice with the PBS, and centrifuged at 2,000 rpm for 5 min to get the required pellet of macrophages.
2.11 Cell culture
The normal human lymphocytes and macrophages, KB and KG1a cell lines, were cultivated for in vitro experiments. Cell lines were obtained from the National Centre for Cell Sciences, Pune, India. It was cultured in RPMI 1640 medium and MEM supplemented with 10 % fetal calf serum, 100 units/ml penicillin and 100 μg/ml streptomycin, 4 mM l-glutamine under 5 % CO2, and 95 % humidified atmosphere at 37 °C.
2.12 Preparation of drug
Several doses of FA-PMIDA-CoO NPs (1–25 μg/ml) were prepared using sterile phosphate-buffered saline (pH 7.4). In this study, all these doses were charged against normal and cancer cell line for evaluation of in vitro anticancer activity.
2.13 In vitro cytotoxicity assay
Each type of cells was divided into nine groups. Each group contained six Petri dishes (2 × 105 cells in each). The cells of each Petri dish of control and experimental groups were maintained in MEM and RPMI 1640 media, where it is applicable, supplemented with 10 % FBS, 50 μg/ml gentamycin, 50 μg/ml penicillin, and 50 μg/ml streptomycin at 37 °C in a 95 % air/5 % CO2 atmosphere in CO2 incubator. From 105 cells/ml cell suspension, 180 ml cell suspension was seeded into each well of 96-well tissue culture plates and incubated for 24 h followed by the addition of FA-PMIDA-CoO NPs, DOX-FA-PMIDA-CoO NPs, and MTX-FA-PMIDA-CoO NPs at concentrations from 1 to 25 μg/ml and was incubated for 24 h at 37 °C in a humidified incubator maintained with 5 % CO2. The cell proliferation was estimated by MTT assay (Chattopadhyay et al. 2012).
2.14 Intracellular uptake study
Nanoparticle uptake by KB and KG1a cells was studied by fluorescence microscopy methods according to our previous laboratory report (Sahu et al. 2010). Cells (2 × 105) were seeded into 35 mm cell culture plates and were incubated with 25 μg/ml FA-PMIDA-CoO-RhB/DOX-FA-PMIDA-CoO-RhB/MTX-FA-PMIDA-CoO-RhB for fluorescence microscopy. Then, the cells were allowed to adhere to a glass coverslip followed by incubation for 4 h at 37 °C in a humidified incubator maintained with 5 % CO2. Fluorescence images were acquired with 540 nm laser for differential interference contrast microscopy and 625 nm lasers for RhB excitation and emission on an Olympus research phase contrast with fluorescence microscope (model CX40; Olympus).
2.15 Interaction of particles with macrophages in vitro
Macrophages isolated from normal human blood were cultured in complete RPMI media and allowed to adhere to the Petri plate surface. Macrophages (5 × 105) in 2 ml complete RPMI media and 50 μg of PMIDA-CoO NP-RhB/FA-PMIDA-CoO NP-RhB were added and incubated for 12 h at 37 °C in a humidified incubator. After incubation, the Petri plates were washed with warm PBS and phagocytic uptake was quantified using phase-contrast fluorescence inverted microscope at ×400 magnifications (Das et al. 2008).
2.16 Apoptosis study by conventional EtBr staining
Apoptosis study was done according to Ribble et al. (2005), briefly suspension cells were harvested and transferred to a 15-ml tube with Dulbecco’s phosphate-buffered saline. Cells were pelleted by centrifugation at 1,000 rpm for 5 min and washed with 1 ml of cold PBS once. Cell pellets were then resuspended in 100 μl cold PBS and 5 μl EtBr dye was added. Stained cell suspension (10 μl) was placed on a clean microscope slide and covered with a coverslip. The cells were viewed and counted using a phase-contrast fluorescence inverted microscope at ×400 magnification with excitation filter 480/30 nm. Pictures were taken with a Nikon COOLPIX digital camera. Tests were done in triplicate, counting a minimum of 100 total cells each.
2.17 Detection of cellular apoptosis by flow cytometric analysis using propidium iodide
Cell apoptosis was measured using propidium iodide staining and analysis by flow cytometry (Roa et al. 2009). After the treatment schedule, the cells were scraped and centrifuged with the supernatant medium at 3,500 rpm for 5 min. Following washes, cells were resuspended in PBS and fixed in 70 % ethanol for 1 h on ice. Fixed cells were washed with PBS and stained with propidium iodide (5 μg/ml) solution containing ribonuclease (RNase) (50 μmol/L) for 30 min at room temperature. Then, the cells were analyzed on a Becton Dickinson FACSCalibur flow cytometer. The G0/G1 population (apoptotic cells) of the cells was determined by CellQuest software.
2.18 Statistical analysis
The data were expressed as mean ± SEM (n = 6). Comparisons between the means of control and treated group were made by one-way ANOVA test (using a Statistical Package, Origin 6.1, Northampton, MA, 01060, USA) with p < 0.05 as a limit of significance.
3 Result and discussion
3.1 Synthesis of FA-PMIDA-CoO nanoparticles
FA-PMIDA-CoO NPs were synthesized according to the method outlined in Scheme 1. –NH2 group of folic acid and carboxylic group (−COOH) of PMIDA-CoO NPs were connected through the end-amino groups. Folic acid was chosen as a linker in order (1) to get optimized length of linker to reach accessible receptor sites, (2) to increase the water solubility, and (3) to make the folate conjugate system biocompatible.
3.2 Solubility and stability assessment
PMIDA-CoO NPs are insoluble in water, whereas FA-PMIDA-CoO NPs form a semitransparent salutation with water even at different pH. The stability of FA-PMIDA-CoO NPs at different pH was estimated spectrophotometrically at 270 nm. Our result shows that FA-PMIDA-CoO NPs shows a very good solubility at pH 2.0 to pH 8.0. PMIDA-CoO NPs are strongly hydrophobic in nature and our previous studies had focused on the biological activities of this water-insoluble particle (Chattopadhyay et al. 2012). To overcome this problem, we had to change sample preparation approaches. We use folic acid as a ligand on PMIDA-coated CoO NPs. The presence of folic acid on outer surface results in water solubility of the metal NPs by visual observation. Solubility in aqueous medium may be advantageous for potential applications of metal NPs in catalysis or biomedicine (Brunner et al. 2006; Uzun et al. 2008). The stability of FA-PMIDA-CoO NPs at different pH was estimated spectrophotometrically at 270 nm. Our result shows that FA-PMIDA-CoO NPs show a very good stability at physiological pH. Amide group is more of a polarizing group than acid group. When amide compound is dissolve in water, it fully coordinates with water through “−H” bonding via “N” atom. As a result, it is very much soluble in water. Each terminal –NH2 group also forms –H bond with water. So, the compound becomes soluble in water (Fig. 1a).
Water solubility of only PMIDA-cobalt oxide nanoparticles and FA-PMIDA-cobalt oxide nanoparticles (a); size determination of FA-PMIDA-cobalt oxide nanoparticles (b), DOX-FA-PMIDA-cobalt oxide nanoparticles (c), and MTX-FA-PMIDA-cobalt oxide nanoparticles (d) by dynamic light scattering. SEM images of PMIDA-coated cobalt oxide nanoparticles (e)
3.3 Dynamic light scattering experiment
The size distribution of hydrophilic FA-PMIDA-CoO NPs in aqueous medium was characterized by DLS. The mean size of nanoparticles in aqueous solution was determined, which ranges from 200 to 240 nm as shown in Fig. 1b. An investigation on hydrodynamic size of drug conjugates −MTX and −DOX was carried out through dynamic light scattering. DOX-FA-PMIDA-CoO shows presence of stable nano-aggregated particles with a hydrodynamic size of 230 ± 30 nm (Fig. 1c) and MTX-FA-PMIDA-CoO of 225 ± 30 nm (Fig. 1d). The stability of drug-conjugated particles was investigated by measuring the hydrodynamic size which shows that there is almost no change of hydrodynamic size. This observation implies that such stable drug conjugates can be circulated in the bloodstream for a long period.
3.4 SEM morphology of FA-PMIDA-CoO nanoparticles
The SEM morphology of FA-PMIDA-CoO NPs shows having nearly spherical geometry with a mean size of less than 100 nm. The result was represented in Fig. 1e. The presence of some bigger particle should be attributed to be aggregating or overlapping of some small particles. The observed NP size was approximately larger than the hydrodynamic diameter obtained from the DLS experiment. SEM imaging describes the size in the dried state of the sample, whereas DLS measured the size in the hydrated state of the sample. So, the size measured by DLS was a hydrodynamic diameter and always larger than the SEM image. However, one has to bear in mind that by SEM, we image single particles, while DLS gives an average size estimation, which is biased toward the larger size end of the population distribution.
3.5 Fourier transform infrared spectroscopy
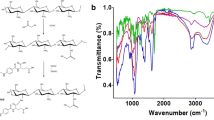
Conjugation of folic acid with PMIDA-coated CoO NPs is investigated by FTIR spectroscopy. The FTIR spectrum of folic acid derivative shows the various functional groups of the molecules. The FTIR spectrum of pure CoO NP, PMIDA, and PMIDA-coated CoO NPs were published in our previous lab report (Chattopadhyay et al. 2012). The comparisons of FTIR spectrum of folic acid (Fig. 2a) and FA-PMIDA-CoO NPs are shown in Fig. 2b. FTIR spectra show basic IR band characteristics of CoO at 566 cm−1, which indicate the presence of CoO vibration and a broad band around 3,440 cm−1, indicative of the presence of –OH groups on the nanoparticle surface. The important areas of examination are in the 3,000–1,500-cm−1 region. This region is known as the functional group region. In FA-PMIDA-CoO, NPs show the characteristic band of folic acid. After conjugation of PMIDA-CoO NPs with folic acid, the spectrum of resultant molecules shows not only the characteristic band of the original band of CoO NPs and PMIDA-CoO NPs, but also the characteristic peak of the folic acid at 1,653 cm−1 (−CONH band) and 1,023 cm−1 (−NH band). Furthermore, the absorption of amide band at 1,023 cm−1 increases, which may be due to the formation of the amide linkage between the amino group of the folic acid and PMIDA-CoO NPs (Fig. 2b). The FTIR spectra of the DOX (ESM 1) particles show strong absorption at 500–600 cm−1, characteristic vibration of M–O in the ferrite lattice (Fig. 2c). In addition to this, the intensification of band at 2,912 cm−1 along with the C=O stretching at 1,645 cm−1 indicates that MTX has been conjugated to –NH2-terminated particles through –CONH linkage. Similarly, the successful attachment of MTX (ESM 1) was also established using FTIR (Fig. 2d).
3.6 In vitro drug release assay
In vitro drug release experiments carried out with MTX and DOX-conjugated NPs in PBS buffer medium at pH 3, pH 5, and pH 7.4 were investigated in two separate batches. Both MTX and DOX showed typical release profiles. MTX showed rapid release rates from the beginning followed by a sustained release pattern of 94 % (pH 3) and 83 % (pH 5), and in PBS (pH 7.4), it was 71 % after 48 h (Fig. 2e). But in the case of DOX, a different release behavior was observed. It was observed that the release of DOX from FA-PMIDA-CoO-DOX NPs in the buffer medium of PBS was 78 % (pH 3) and 58 % (pH 5), whereas in PBS (pH 7.4), it was 45 % after 48 h. The overall release rates were higher and lower pH (7.0 and 3.0) and less release was observed in physiological pH. This is because the drugs have been covalently grafted through amide bonds and protonation of amino groups at lower pH which loosens nanoparticle structure (Sahu et al. 2010). These pH-sensitive cleavages of amide bonds release the drug (Fig. 2e). The extracellular pH of tumors is slightly more acidic than the blood and normal tissue (Engin et al. 1995; Van Sluis et al. 1999). It was observed that at acidic pH (pH 5.0), MTX and DOX release was more than physiological pH. In addition, it is proposed that nanoparticles are internalized by cells via an endocytosis process (Park et al. 2006; Decuzzi and Ferrari 2007). The endocytic pathway begins near the physiological pH of 7.4; it drops to a lower pH of 5.5–6.0 in endosomes and approaches pH 4.5–5.0 in lysosomes (Mellman et al. 1986). Therefore, polymeric nanoparticles that are responsive to pH can be designed to selectively release their payload in tumor tissue or within tumor cells.
3.7 Plasma protein binding activity
The surface chemistry of biomaterials has great effects on the protein adsorption process. Some factors such as electrostatic interaction, hydrophobic interaction, and specific chemical interactions between the protein and the adsorbent play important roles. The binding of proteins to FA-CoO NP is shown in Fig. 3b, c. Our result supported that NPs can bind many different plasma proteins. The knowledge of adsorption of albumin to the NPs is very important because once in the body, blood proteins will adsorb to the particles and cells will then react with the adsorbed proteins on the particles which will ultimately affect cellular uptake and can alter biochemical activity.
3.8 In vitro toxicity studies
The toxicity of the FA-PMIDA-CoO NPs is checked towards normal human lymphocytes in vitro. FA-PMIDA-CoO NP-mediated cytotoxicity to these normal cells was measured by MTT assay (Fig. 4a). Here, we investigated the effect of FA-PMIDA-CoO NPs, FA-PMIDA-CoO-DOX NPs, and FA-PMIDA-CoO-MTX NPs on the proliferation of FR (+) KB cells and FR (−) KG1a cells in vitro. It is observed from our experiment that PMIDA-coated CoO NPs kill the KB cells by 12.20, 23.58, 35.78, and 58.18 and KG1a cells by 11.58, 22.36, 34.82, and 55.27 %, respectively, at a dose of 1, 5, 10, and 25 μg/ml (significant at the level of p < 0.05) (Fig. 4b). Whereas FA-PMIDA-CoO NPs are nontoxic at the dose of 1, 5, 10, and 25 μg/ml (Fig. 4a). So, it is observed from our results that FA-PMIDA-CoO NPs have no effect on KB and KG1a cell lines as well as normal human immunocytes.
Cytotoxicity of FA-PMIDA-CoO nanoparticles towards normal human immune cells (d), cytotoxicity of PMIDA-CoO nanoparticles and FA-PMIDA-CoO nanoparticles towards KB and KG1a cell lines (a), and cytotoxicity of DOX-FA-PMIDA-CoO nanoparticles and MTX-FA-PMIDA-CoO nanoparticles towards KB and KG1a cell lines (b). n = 6; values are expressed as mean ± SEM. Asterisk indicates the significant difference as compared to control group
It was observed that DOX-FA-PMIDA-CoO NPs and MTX-FA-PMIDA-CoO NPs induce a significant change in the proliferation with a concentration up to 25 μg/ml with respect to the control, suggesting FA-PMIDA-CoO NPs as an efficient drug carrier. Subsequently, the proliferation of KB and KG1a cells reduced significantly in the presence of DOX-FA-PMIDA-CoO NPs and MTX-FA-PMIDA-CoO NPs at 1–25 μg/ml in respect to FA-PMIDA-CoO NPs alone. This is possibly attributable to the affinity of folic acid towards the folate receptor, which results in higher intake of DOX-FA-PMIDA-CoO and MTX-FA-PMIDA-CoO particles as compared to FA-PMIDA-CoO NPs. However, the proliferation of KB and KG1a cells reduced drastically in a dose-dependent manner in the presence of drug-conjugated NPs. This is attributed to the active uptake of nanoparticles in a folate receptor-mediated endocytosis that interfered with the cell proliferation, which ultimately results in the target-specific delivery of drugs to cancer cells (Fig. 4c). DOX-FA-PMIDA-CoO particles kill KB cells by 28.06, 55.27, 75.43, and 85.16 % and KG1a cells by 15.83, 26.71, 36.63, and 47.71 %; MTX-FA-PMIDA-CoO kills KB cells by 27.92, 52.64, 72.86, and 85.29 % and KG1a cells by 14.54, 25.29, 36.2, and 48.65 % at the dose of 1–25 μg/ml.
3.9 Intracellular uptake by fluorescence imaging
Nanoparticles binding to the plasma membrane and cellular uptake are probably a necessary condition for its exertion of cytotoxicity. We have shown that drug-loaded FA-PMIDA-CoO NPs were internalized by cancer cells in in vitro cultures and the alteration of cellular structure was observed (Fig. 5a, d). This internalization might be due to the receptor-mediated endocytosis (Mohapatra et al. 2007). KB cells express greater amount of FR and the internalization of drug-conjugated NPs was higher, but KG-1A cells express lower FR on cell surfaces and internalization of drug-conjugated NPs was lower than the KB cells. For this reason, KB cells are more susceptible to drug-conjugated NPs than KG-1A cells.
3.10 Phagocytosis of particles by macrophages in vitro
Macrophages are the primary phagocytes in the body, which play a critical role in the clearance of particles delivered into the body’s circulation. The phagocytosis of FA-PMIDA-CoO NPs by macrophages was studied in vitro. It was observed that phagocytosis of the folate-adorned nanoparticles by macrophages was appreciably lower, as compared to the uptake by KB cells (Fig. 5e, f). The uptake level is even higher when PMIDA-CoO NPs have been used (Fig. 5e). It should, however, be noted that, with the present size and surface chemistry of nanoparticles, the phagocytosis by macrophages will be low compared to folate receptor-mediated endocytosis by cancer cells. The present finding is also consistent with an earlier report on the phagocytosis of folate-modified nanoparticles (Das et al. 2008). FR is known to be upregulated in a cancer cells, whereas in normal cells, FR is only minimally distributed (Kamen and Smith 2004). Our result showed that higher amount of FA-PMIDA-CoO NPs was internalized into the KB cells and lower amount into FR lower expressing cells. For that reason, FA-PMIDA-CoO NPs did not produce any adverse effects towards normal cells up to selected dose.
3.11 Cellular apoptosis analysis by EtBr staining and flow cytometric study
To investigate whether FA-PMIDA-CoO NPs are associated with the induction of cellular apoptosis, after the treatment schedule, the phenotypic characteristics of cells were evaluated by fluorescence microscopic examination of overall morphology at a magnification of ×50. Ethidium bromide was used to visualize condensed chromatin of apoptotic dead cells. The treated cells showed red color because ethidium bromide stains fragmented nuclear chromatin in apoptotic cells (Fig. 6), but it is excluded by live cells (Meiyanto et al. 2007).
Fluorescence image of the EtBr-stained KB after DOX-FA-PMIDA-CoO (a, b) and MTX-FA-PMIDA-CoO (c, d) and the image of the EtBr-stained KG1a cells after DOX-FA-PMIDA-CoO (e, f) and MTX-FA-PMIDA-CoO (g, h). Each of the figures mentioned here was represented in gray scale and fluorescence scale, respectively
A quantitative evaluation of DOX-FA-PMIDA-CoO NP- and MTX-FA-PMIDA-CoO NP-mediated cell death was carried out through cell cycle analysis using standard PI staining flow cytometry. Figure 7 shows an increase in cells in the sub-G0/G1 phase (P1) with an increase in fragmented DNA. The anticancer effect of drug-loaded CoO NPs is due to increasing chromatin condensation and DNA fragmentation. DNA fragmentation in two cell lines can be seen on the results of staining with PI, which indicates the occurrence of apoptosis (Fig. 7), and characteristic features of chromatin condensation and fragmentation of DNA (Qu et al. 2007). DOX and MTX are two potential anticancer drugs. Therefore, we intended to study whether the targeted delivery of DOX and MTX in a folate receptor-mediated method could result in a similar cell fate. Hence, we investigated the nuclear fragmentation which is a hallmark of apoptosis with the cells treated with DOX- and MTX-loaded particles. With an increasing dose of the DOX-FA-PMIDA-CoO NPs and MTX-FA-PMIDA-CoO NPs, an increase in the formation of fragmented nuclei containing condensed nuclear material was observed in fluorescence microscopy after nuclear staining with EtBr.
DOX-FA-PMIDA-CoO NP-induced cellular apoptosis analysis in KB and KG1a cell lines by flow cytometry using propidium iodide. Here, a (KB cells) = control, b and c = KB cell + 10 and 25 μg/ml DOX-FA-PMIDA-CoO, respectively, d and e = KB cells + 10 and 25 μg/ml MTX-FA-PMIDA-CoO, respectively; nanoparticles induced cellular apoptosis analysis KG1a cell lines by flow cytometry using propidium iodide. Here, f = control, g and h = KG1a cell + 10 and 25 μg/ml DOX-FA-PMIDA-CoO, respectively, i and j = KG1a cells + 10 and 25 μg/ml MTX-FA-PMIDA-CoO, respectively. Here, P1 = G0/G1 phase (apoptotic phase), P2 = S phase (synthesis phase), P3 = M phase
4 Conclusion
In this study, a simple process has been developed to synthesize FA-PMIDA-CoO NPs. This nano polymeric carrier system shows excellent solubility in aqueous medium with reasonable good hydrodynamic size. In vitro cytotoxicity test using an MTT assay method showed that these FA-PMIDA-CoO NPs have no adverse effect on normal cells. Surface functionalization with –COOH and –NH2 groups facilitates conjugation of nanoparticles with anticancer drugs through amide which is efficiently uptaken by cancer cells. The data presented here suggest that further in vivo studies are warranted to define the therapeutic index of the tumor-targeting polymeric carrier and will constitute the basis for the next-generation drug development.
Abbreviations
- CoO:
-
Cobalt oxide
- DOX:
-
Doxorubicin
- EDC:
-
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide
- EtBr:
-
Ethidium bromide
- FA:
-
Folic acid
- FACS:
-
Flow cytometry
- FR:
-
Folic receptor
- HCl:
-
Hydrochloric acid
- K2HPO4:
-
Dipotassium hydrogen phosphate
- KH2PO4:
-
Potassium dihydrogen phosphate
- M:
-
Molar concentration
- MEM:
-
Minimum essential medium
- MTT:
-
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- MTX:
-
Methotrexate
- NHS:
-
N-hydroxysuccinimide
- NP:
-
Nanoparticles
- PMIDA:
-
Phosphonomethyl iminodiacetic acid
- PBMC:
-
Peripheral blood mononuclear cells
- RhB:
-
Rhodamine B
References
Antony AC (1996) Folate receptors. Annu Rev Nutr 16:501–521. doi:10.1146/annurev.nu.16.070196.002441
Brunner TJ, Wick P, Manser P, Spohn P, Grass RN, Limbach LK, Bruinink A, Stark WJ (2006) In vitro cytotoxicity of oxide nanoparticles: comparison to asbestos, silica, and the effect of particle solubility. Environ Sci Technol 40:4374–4381. doi:10.1021/es052069i
Chakraborty SP, Sahu SK, KarMahapatra S, Santra S, Bal M, Roy S, Pramanik P (2010) Nanoconjugate vancomycin: new opportunities for the development of anti-VRSA agents. Nanotechnology 21:105103. doi:10.1088/0957-4484/21/10/105103
Chattopadhyay S, Chakraborty SP, Laha D, Baral R, Pramanik P, Roy S (2012) Surface-modified cobalt oxide nanoparticles: new opportunities for anti-cancer drug development. Cancer Nano. doi:10.1007/s12645-012-0026-z
Das M, Mishra D, Maiti TK, Basak A, Pramanik P (2008) Bio-functionalization of magnetite nanoparticles using an aminophosphonic acid coupling agent: new, ultradispersed, iron-oxide folate nanoconjugates for cancer-specific targeting. Nanotechnology 19:415101. doi:10.1088/0957-4484/19/41/415101
Decuzzi P, Ferrari M (2007) The role of specific and non-specific interactions in receptor-mediated endocytosis of nanoparticles. Biomaterials 28:2915–2922
Elnakat H, Ratnam M (2004) Distribution, functionality and gene regulation of folate receptor isoforms: implications in targeted therapy. Adv Drug Deliv Rev 56:1067–1084. doi:10.1016/j.addr.2004.01.001
Engin K, Leeper DB, Cater JR, Thistlethwaite AJ, Tupchong L, McFarlane JD (1995) Extracellular pH distribution in human tumors. Int J Hyperth 11:211–216
Faraji AH, Wipf P (2009) Nanoparticles in cellular drug delivery. Bioorg Med Chem 17:2950–2962. doi:10.1016/j.bmc.2009.02.043
Ghosh T, Chattopadhyay T, Das S, Mondal S, Suresh E, Zangrando E, Das D (2011) Thiocyanate and dicyanamide anion controlled nuclearity in Mn, Co, Ni, Cu, and Zn metal complexes with hemilabile ligand 2-benzoylpyridine. Cryst Growth Des 11:3198–3205. doi:10.1021/cg2004485
Hudson L, Hay FC (1991) Practical immunology, 3rd ed. Blackwell, Oxford, pp 21–22
Kamen BA, Capdevila A (1986) Receptor-mediated folate accumulation is regulated by the cellular folate content. Proc Natl Acad Sci 83:5983–5987, PMCID: PMC386421
Kamen BA, Smith AK (2004) A review of folate receptor alpha cycling and 5-methyltetrahydrofolate accumulation with an emphasis on cell models in vitro. Adv Drug Deliv Rev 56:1085–1097. doi:10.1016/j.addr.2004.01.002
Leamon CP, Low PS (1991) Delivery of macromolecules into living cells: a method that exploits folate receptor endocytosis. Proc Natl Acad Sci 88:5572–5576, PMCID: PMC51919
Leamon CP, Reddy JA (2004) Folate-targeted chemotherapy. Adv Drug Deliv Rev 56:1127–1141. doi:10.1016/j.addr.2004.01.008
Lee RJ, Low PS (1994) Delivery of liposomes into cultured KB cells via folate receptor-mediated endocytosis. JBC 269(5):3198–3204
Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, Tamanoi F, Zink JI (2008) Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano 2:889–896. doi:10.1021/nn800072t
Low PS, Antony AC (2004) Folate receptor-targeted drugs for cancer and inflammatory diseases. Adv Drug Deliv Rev 56:1057–1238. doi:10.1016/j.addr.2004.02.003
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275, PMID:14907713
Maeda H, Bharate GY, Daruwalla J (2009) Polymeric drugs for efficient tumor-targeted drug delivery based on EPR-effect. Eur J Pharm Biopharm 71:409–419. doi:10.1016/j.ejpb.2008.11.010
Marin RV, Ng CH, Wilke M, Tiersch B, Fratzl P, Peter MG (2005) Size controlled hydroxyapatite nanoparticles as self-organized organic-inorganic composite materials. Biomaterials 26:5414–5426. doi:10.1016/j.biomaterials.2005.01.051
Meiyanto E, Agustina D, Suparjan AM, Da’ IM (2007) PVG-O induces apoptosis on T47D breast cancer cells line through caspase-3 activation. Jurnal Kedokteran Yarsi 15:075–079
Mellman I, Fuchs R, Helenius A (1986) Acidification of the endocytic and exocytic pathways. Annu Rev Biochem 55:773–800
Mohapatra S, Mallick SK, Maiti TK, Ghosh SK, Pramanik P (2007) Synthesis of highly stable folic acid conjugated magnetite nanoparticles for targeting cancer cells. Nanotechnology 18:385102–385111. doi:10.1088/0957-4484/18/38/385102
Mohapatra S, Rout SR, Maiti S, Maiti TK, Panda AB (2011) Monodisperse mesoporous cobalt ferrite nanoparticles: synthesis and application in targeted delivery of antitumor drugs. J Mater Chem 21:9185. doi:10.1039/C1JM10732A
Pan XQ, Zheng X, Shi G, Wang H, Ratnam H, Lee RJ (2002) A strategy for the treatment of acute myelogenous leukemia based on folate receptor type b-targeted liposomal doxorubicin in combined with receptor induction using all-trans retinoic acid. Blood 100:594–602. doi:10.1182/blood.V100.2.594
Panyam J, Labhasetwar V (2003) Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev 55:329–347. doi:10.1016/S0169-409X(02)00228-4
Papis E, Rossi F, Raspanti M, Isabella DD, Colombo G, Milzani A, Bernardini G, Gornati R (2009) Engineered cobalt oxide nanoparticles readily enter cells. Toxicol Lett 189:253–259. doi:10.1016/j.toxlet.2009.06.851
Park JS, Han TH, Lee KY, Han SS, Hwang JJ et al (2006) N-acetyl histidine-conjugated glycol chitosan self-assembled nanoparticles for intracytoplasmic delivery of drugs: endocytosis, exocytosis and drug release. J Control Release 115:37–45
Patila S, Sandbergb A, Heckertc E, Self W, Seal S (2007) Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 28(31):4600–4607. doi:10.1016/j.biomaterials.2007.07.029
Qu X, Cui S, Tian Z, Li X, Chen M, Xu FW, Inagaki Y, Deng BY, Makuuchi M, Nakata M, Tang W (2007) Induction of apoptosis in human hepatocellular carcinoma cells by synthetic antineoplaston A10. Anticancer Res 27:2427–2432, PMID:17695534
Ribble D, Goldstein NB, Norris AD, Shellman YG (2005) A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol 5:12. doi:10.1186/1472-6750-5-12
Roa W, Xiaojing Z, Guo L, Shaw A, Hu X, Xiong Y, Gulavita S, Patel S, Sun X, Chen J, Moor R, Xing JZ (2009) Gold nanoparticles sensitize radiotherapy of prostate cancer cell by regulation of the cell cycle. Nanotechnology 20:1–9. doi:10.1088/0957-4484/20/37/375101
Sahu SK, Mallick SK, Santra S, Maiti TK, Ghosh SK, Pramanik P (2010) In vitro evaluation of folic acid modified carboxymethyl chitosan nanoparticles loaded with doxorubicin for targeted delivery. J Mater Sci Mater Med 21:1587–1597. doi:10.1007/s10856-010-3998-4
Uzun O, Hu Y, Verma A, Chen S, Centrone A, Stellacci F (2008) Water-soluble amphiphilic gold nanoparticles with structured ligand shells. Chem Commun 2:196–198. doi:10.1039/b713143g
Van Sluis R, Bhujwalla ZM, Ballerteros P, Alverez J, Cerdan S, Galons JP et al (1999) In vivo imaging of extracellular pH using 1H MSRI. Magn Reson Med 41:743–750
Wang K, Xu JJ, Chen HY (2005) A novel glucose biosensor based on the nanoscaled cobalt phthalocyanine-glucose oxidase biocomposite. Biosens Bioelectron 20:1388–1396. doi:10.1016/j.bios.2004.06.006
Acknowledgments
The authors express gratefulness to the Department of Biotechnology, Government of India for funding and to the Indian Institute of Technology, Kharagpur and Vidyasagar University, Midnapore for providing the facilities to execute these studies.
Conflict of interest
The authors declare that there are no conflicts of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Sa and Sb represent the FTIR spectra of only doxorubicin and methotrexate, respectively (DOC 646 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chattopadhyay, S., Dash, S.K., Ghosh, T. et al. Surface modification of cobalt oxide nanoparticles using phosphonomethyl iminodiacetic acid followed by folic acid: a biocompatible vehicle for targeted anticancer drug delivery. Cancer Nano 4, 103–116 (2013). https://doi.org/10.1007/s12645-013-0042-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12645-013-0042-7