Abstract
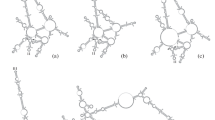
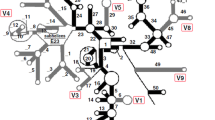
Previously, it has been found that species of the zoanthid genus Zoanthus (Cnidaria: Hexacorallia) possess high levels of intrageneric ITS-rDNA sequence variation both between congeners and also within one species, Zoanthus sansibaricus, resulting in an uncertain internal transcribed spacer of ribosomal DNA (ITS-rDNA) phylogeny for this group. For the first time, the secondary structures of the internal transcribed spacer 2 (ITS 2) sequences were analyzed in an attempt to further clarify and solve relationships within Zoanthus, and also with the closely related genera Acrozoanthus and Isaurus (all in family Zoanthidae). Results show that most species’ ITS2 secondary structures follow the basic four-helix-ring model proposed for most eukaryotes including anthozoans. However, not all structures had the conserved motifs present in scleractinian corals, and Z. sansibaricus had two different structural conformations from the two different ITS2 types present. The ITS2 secondary structures of Zoanthus in this study, present a well resolved and supported phylogeny that makes it an appropriate tool for solving phylogenies in this taxonomic group. Based on ITS2 secondary structure results here, we theorize that: (1) Acrozoanthus is a valid monophyly separate from Zoanthus; (2) Z. gigantus, Z. sansibaricus, and Z. praelongus do not have any compensatory base changes (CBCs) between them and form a related clade distinct from the more divergent Z. kuroshio/Z. vietnamensis clade. The results furthermore suggest that the presence of CBCs may exist only at much lower ratios between different species in zoanthids than in other investigated organism lineages.


Similar content being viewed by others
References
Aguilar C, Sánchez JA (2007) Phylogenetic hypothesis of gorgoniid octocorals according to ITS2 and their predicted RNA secondary structures. Mol Phylogenet Evol 43:774–786
Ahvenniemi P, Wolf M, Lehtonen MJ, Wilson P, German-Kinnari M, Valkonen JPT (2009) Evolutionary diversification indicated by compensatory base changes in ITS2 secondary structures in a complex fungal species, Rhizoctonia solani. J Mol Evol 69:150–163
Berntson EA, Bayer FM, McArthur AG, France SG (2001) Phylogenetic relationships within the Octocorallia (Cnidaria: Anthozoa) based on nuclear 18 S rRNA sequences. Mar Biol 138:235–246
Carlgren O (1954) Actiniaria and Zoantharia from South and West Australia with comments upon some Actiniaria from New Zealand. Arkiv för Zoologi (2) 6:571–595
Chen CA, Chang CC, Wei NV, Chen CH, Lein YT, Lin HE, Dai CF, Wallace CC (2004) Secondary structure and phylogenetic utility of the ribosomal internal transcribed spacer 2 (ITS2) in scleractinian corals. Zool Stud 43:759–771
Coleman AW, van Oppen MJH (2008) Secondary structure of the rRNA ITS2 region reveals key evolutionary patterns in acroporid corals. J Mol Evol 67:389–396
Côte CA, Greer CL, Peculis BA (2002) Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA (N Y) 8:786–797
Elder JF Jr, Turner BJ (1995) Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol 70:297–320
Fabry S, Kohler A, Coleman AW (1999) Intraspecies analysis: comparison of ITS sequence data and gene intron sequence data with breeding data for a worldwide collection of Gonium pectorale. J Mol Evol 48:94–101
Grajales A, Aguilar C, Sánchez JA (2007) Phylogenetic reconstruction using secondary structures of internal transcribed spacer 2 (ITS2, rDNA): finding the molecular and morphological gap in Caribbean gorgonian corals. BMC Evol Biol 7:90
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Haddon AC, Shackleton AM (1891) Reports on the zoological collections made in Torres Straits by Professor A.C. Haddon, 1888–1889. Actiniae: I. Zoantheae. Sci Trans R Dublin Soc 4:673–701
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Han K, Yanga B (2003) PseudoViewer2: visualization of RNA pseudoknots of any type. Nucleic Acids Res 31:3432–3440
Harris DJ, Crandall KA (2000) Intragenomic variation within ITS1 and ITS2 of freshwater crayfishes (Decapoda: Cambaridae): Implications for phylogenetic and microsatellite studies. Mol Biol Evol 17:284–291
Hillis DM, Dixon MT (1991) Ribosomal DNA: molecular evolution and phylogenetic inference. Quart Rev Biol 66:411–453
Huang D, Meier R, Todd PA, Chou LM (2008) Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J Mol Evol 66:167–174
Katoh K, Hiroyuki T (2008) Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinform 9:212
Keller A, Schleicher T, Schultz J, Mullar T, Dandekar T, Wolf M (2009) 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene 430:50–57
Keller A, Forster F, Mullar T, Dandekar T, Schultz J, Wolf M (2010) Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol Direct 4(5):1–12
Kishino H, Hasegawa H (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179
Koehl MAR (1977) Water flow and the morphology of zoanthid colonies. Proc 3rd Int Coral Reef Symp 1:437–444
Muirhead A, Ryland JS (1985) A review of the genus Isaurus Gray 1828 (Zoanthidea), including new records from Fiji. J Nat Hist 19:323–335
Müller T, Philippi N, Dandekar T, Schultz J, Wolf M (2007) Distinguishing species. RNA (N Y) 13:1469–1472
Odonnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
Oliverio M, Barco A, Modica MV, Richter A, Mariottini P (2009) Ecological barcoding of corallivory by second internal transcribed spacer sequences: hosts of coralliophiline gastropods detected by the cnidarian DNA in their stomach. Mol Ecol Resour 9:94–103
Pax F, Mueller I (1957) Zoantharien aus Viet-Nam. Mem Mus Natl Hist Nat 16:1–40
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Reimer JD, Ono S, Takishita K, Fujiwara Y, Tsukahara J (2004) Reconsidering Zoanthus spp. diversity: molecular evidence of conspecifity within four previously presumed species. Zool Sci 21:517–525
Reimer JD, Ono S, Iwama A, Tsukahara J, Maruyama T (2006a) High levels of morphological variation despite close genetic relatedness between Zoanthus aff. vietnamensis and Zoanthus kuroshio (Anthozoa: Hexacorallia). Zool Sci 23:755–761
Reimer JD, Ono S, Iwama A, Tsukahara J, Takishita K, Maruyama T (2006b) Morphological and molecular revision of Zoanthus (Anthozoa: Hexacorallia) from southwestern Japan with description of two new species. Zool Sci 23:261–275
Reimer JD, Ono S, Takishita K, Tsukahara J, Maruyama T (2006c) Molecular evidence suggesting species in the zoanthid genera Palythoa and Protopalythoa (Anthozoa: Hexacorallia) are congeneric. Zool Sci 23:87–94
Reimer JD, Takishita K, Ono S, Maruyama T (2007a) Diversity and evolution in the zoanthid genus Palythoa (Cnidaria: Hexacorallia) utilizing nuclear ITS-rDNA. Coral Reefs 26:399–410
Reimer JD, Takishita K, Ono S, Tsukahara J, Maruyama T (2007b) Molecular evidence suggesting intraspecific hybridization in Zoanthus (Anthozoa: Hexacorallia). Zool Sci 24:346–359
Reimer JD, Ono S, Tsukahara J, Iwase F (2008) Molecular characterization of the zoanthid genus Isaurus (Anthozoa: Hexacorallia) and its zooxanthellae (Symbiodinium spp). Mar Biol 153:351–363
Ronquist F, Huelsenbeck JP (2003) MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxf) 19:1572–1574
Sánchez JA, Dorado D (2008) Intragenomic ITS2 variation in Caribbean seafans. Proc 11th Int Coral Reef Symp 7–11
Schultz J, Maisel S, Gerlach D, Müller T, Wolf M (2005) A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA (N Y) 11:361–364
Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M (2006) 4SALE-A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform 7:498
Shearer TL, van Oppen MJH, Romano SL, Wörheide G (2002) Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol Ecol 11:2475–2487
Sinniger F, Montoya-Burgess JI, Chevaldonne P, Pawlowski J (2005) Phylogeny of the order Zoantharia (Anthozoa, Hexacorallia) based on mitochondrial ribosomal genes. Mar Biol 147:1121–1128
Sinniger F, Reimer JD, Pawlowski J (2008) Potential of DNA sequences to identify zoanthids (Cnidaria: Zoantharia). Zool Sci 25:1253–1260
Swofford DL (1998) PAUP*. V Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer, Sunderland
Vollmer SV, Palumbi SR (2004) Testing the utility of internally transcribed spacer saequences in coral phylogenetics. Mol Ecol 13:2763–2772
Wei NWV, Wallace CC, Dai CF, Pillay KRM, Chen CA (2006) Analyses of the ribosomal internal transcribed spacers (ITS) and the 5.8 S gene indicate that extremely high rDNA heterogeneity is a unique feature in the scleractinian coral genus Acropora (Scleractinia; Acroporidae). Zool Stud 45:404–418
Wolf M, Achtziger M, Schultz J, Dandekar T, Müller T (2005a) Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA (N Y) 11:1616–1623
Wolf M, Friedrich J, Dandekar T, Müller T (2005b) CBCAnalyzer: inferring phylogenies based on compensatory base changes in RNA secondary structures. In Silico Biol 5:0027
Wolf M, Ruderisch B, Dandekar T, Müller T (2008) ProfdistS: (Profile-) Distance based phylogeny on sequence-structure alignments. Bioinformatics (Oxf) 24:2401–2402
Wörheide G, Nichols SA, Goldberg J (2004) Intragenomic variation of the rRNA internal transcribed spacers in sponges (Phylum Porifera): implications for phylogenetic studies. Mol Phylogenet Evol 33:816–830
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgements
C.A. was supported by the Japanese Government (Monbukagakusho: MEXT) Scholarship in the Faculty of Science at the University of the Ryukyus. We wish to thank Dr. Matthias Wolf (Department of Bioinformatics, University of Würzburg) for his continuous support with software applications. Dr. F. Sinniger funded by the Rectors’ Conference of the Swiss Universities (CRUS) is thanked for Acrozoanthus ITS sequence. J.D.R. was supported by the Rising Star Program at the University of the Ryukyus and a “Wakate B” research grant from the Japan Society for the Promotion of Science. Three anonymous reviewers greatly improved this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 644 kb)
Rights and permissions
About this article
Cite this article
Aguilar, C., Reimer, J.D. Molecular phylogenetic hypotheses of Zoanthus species (Anthozoa:Hexacorallia) using RNA secondary structure of the internal transcribed spacer 2 (ITS2). Mar Biodiv 40, 195–204 (2010). https://doi.org/10.1007/s12526-010-0043-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12526-010-0043-2