Abstract
Background
Transcatheter aortic valve implantation (TAVI) has become a commonly applied procedure for high-risk aortic valve stenosis patients. However, for some patients, this procedure does not result in the expected benefits. Previous studies indicated that it is difficult to predict the beneficial effects for specific patients. We aim to study the accuracy of various traditional machine learning (ML) algorithms in the prediction of TAVI outcomes.
Methods and results
Clinical and laboratory data from 1,478 TAVI patients from a single centre were collected. The outcome measures were improvement of dyspnoea and mortality. Three experiments were performed using (1) screening data, (2) laboratory data, and (3) the combination of both. Five well-established ML techniques were implemented, and the models were evaluated based on the area under the curve (AUC). Random forest classifier achieved the highest AUC (0.70) for predicting mortality. Logistic regression had the highest AUC (0.56) in predicting improvement of dyspnoea.
Conclusions
In our single-centre TAVI population, the tree-based models were slightly more accurate than others in predicting mortality. However, ML models performed poorly in predicting improvement of dyspnoea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
-
This is the first study on the prediction of TAVI outcomes using machine learning (ML) techniques.
-
We have shown that ML techniques slightly outperform traditional methods in predicting mortality in our patient population; traditional logistic modelling has more prognostic value in predicting improvement of dyspnoea.
-
N-terminal pro-b-type natriuretic peptide, body mass index, chronic kidney disease epidemiology collaboration, creatinine, and patient age were the most important features in our ML models.
Introduction
Aortic valve stenosis (AS) is one of the most common valvular heart diseases, impacting, in general, the elderly population. In the past decade, transcatheter aortic valve implantation (TAVI) has developed into a routine treatment for AS patients at elevated risk of surgery. Although there is strict patient selection for the TAVI procedure and various planning and treatment support tools are available [1,2,3], a number of patients have limited benefit from TAVI [4]. Improved selection of these patients would allow increased benefit from the procedure and improve decision-making. Unfortunately, current risk models have only limited accuracy in predicting TAVI outcomes [5].
Previous clinical prediction models rely on traditional statistical regression models [6]. Alternatively, machine learning (ML), which is a computer science subdiscipline, has shown superior predictive value in various clinical areas, from detecting Alzheimer’s disease to identifying lung nodules [7, 8]. A more specific area of ML is supervised learning: with known outcomes, ML algorithms can learn automatically to optimise the prediction of this outcome. Moreover, ML techniques have outperformed conventional regression models when applied to a large amount of data [9].
Multiple risk models that have been used that are dedicated to the prediction of perioperative mortality and are not TAVI-specific, but intended for surgical aortic valve replacement such as the EuroSCORE, EuroSCORE II or the STS (Society of Thoracic Surgery) score [10, 11]. For TAVI, these are poor predictors of mortality and focus on procedural or 30-day mortality, as did the TAVI-specific TVT registry score [12]. The prediction of 1‑year mortality is even more challenging [13]. A more recent study also incorporated predefined features from computed tomography (CT) in combination with comorbidities to enhance the model [14].
We aimed to study the accuracy of various ML algorithms in predicting outcomes after a TAVI procedure. The accuracy was evaluated in the prediction of mortality and improvement of dyspnoea using a subset of well-established ML techniques.
Methods
Patient population
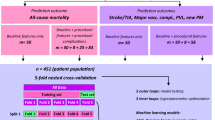
The database consists of 1,478 patients who underwent a TAVI between 2007 and 2018; their median age is 82.9 years [Q1 78.0 − Q3 86.4] and 55% of the patients are female. The data contain patient characteristics, medical history, symptoms, and test results prior to and after TAVI. Symptoms are dyspnoea, fatigue, collapse, and angina pectoris. Tests performed prior to TAVI are echocardiography, computed tomography angiography, coronary angiography, electrocardiography (ECG), and laboratory tests. Tests done after TAVI are echocardiography, ECG and laboratory tests.
The outcomes used are improvement of dyspnoea and mortality. Dyspnoea is measured using the New York Heart Association (NYHA) functional score (1–4). Mortality is defined as a patient who died of a cardiovascular disease within 1 year after the procedure. Patients with missing data are excluded. The baseline and 60-day follow-up NYHA score is known for 766 patients (605 improved, 161 non-improvements) and mortality is known for 1,400 patients (1,263 survivors, 137 non-survivors). For every outcome parameter, we performed three experiments: (1) using screening data; (2) using laboratory data; and (3) using both screening and laboratory data. The number of patients for each experiment is different due to missing values, as presented in Figs. 1 and 2. All variables, as well as the descriptive statistics, can be found in the Electronic Supplementary Material (Tables I and II).
Clinical variables
The clinical variables used can be divided into three sets: patient characteristics, screening data, and laboratory data. The patient baseline characteristics included age, sex, body mass index (BMI), and access route chosen for the procedure. The screening data consist of the medical history, symptoms, and echocardiography prior to TAVI. The features used from this data are the existence of peripheral artery disease, chronic obstructive pulmonary disease (COPD), atrial fibrillation, diabetes mellitus (DM) in the medical history, left ventricular function, and aortic valve area assessed with echocardiography. The features used from the laboratory data are the pre-procedural values of N‑terminal pro-b-type natriuretic peptide (NT-proBNP), haemoglobin, albumin, chronic kidney disease epidemiology collaboration (CKD-EPI), and creatinine.
Based upon expert opinions we applied clipping to the NT-proBNP and creatinine variables, for values greater than 1,000 ng/l and 250 mmol/l, respectively. The nominal and categorical data were one-hot encoded; continuous features were normalised by removing the mean and scaling to unit variance, as requisite for many ML techniques [15]. Moreover, the COPD and DM were dichotomised to take into account the presence of the disease instead of the degree.
Classification techniques
In this study, we selected a number of well-established ML techniques, which are: support vector machine (SVM) [16], random forest classifier (RFC) [17], multi-layer perceptron (MLP) [18], and gradient tree boosting (GTB) [19]. In addition, traditional logistic regression (LR) was also applied for comparison, since this technique is often used in clinical studies. All the implementations used in this project were provided by scikit-learn [15], except for GTB. We chose the XGBoost [19] library because of its GPU implementation, which speeds up training and optimisation.
To evaluate the models fairly, the database was split into two sets: a training and a testing dataset. The training data were used to find the optimal parameters for the classification task. The testing set was used to evaluate the trained model in unseen data, to ensure generalisation of the model and prevent the memorisation of the training set (overfitting). In this study, the models were evaluated with the Monte-Carlo cross-validation for 100 iterations and stratified splits of 70% for training and 30% for testing. With this large number of different training and testing sets, chances of having over-optimistic results are minimised. Moreover, to optimise the parameters of each model, a randomised grid search with stratified 5‑fold cross-validation was performed using the training set. The hyperparameters and ranges used for optimisation, including the weight penalisation applied to minimise the class unbalance issue, are available in the Electronic Supplementary Material (Tables III and IV).
Results of ML techniques are difficult to interpret. To elucidate which features may be important in the ML techniques, the average feature importance for RFC and GTB was calculated based on the number of times the feature was selected for splitting and weighted by the average squared improvement of the model over all trees [20].
Performance assessment
The median of the area under the curve (AUC) of the receiver operating characteristic curve (ROC) from 100 iterations, using test sets, was selected to evaluate the performance of each model. To assess whether the difference in AUC between highest performing classifier and the other methods was statistically significant, the Wilcoxon signed-rank test was performed for each experiment. p-values < 0.05 were considered statistically significant.
Results
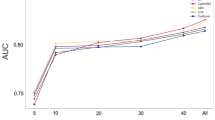
The predictive value for improvement of dyspnoea was statistically significant but absent/low, with the best median AUC result of 0.56, using only laboratory features and LR. For mortality prediction, the model based on RFC was most accurate with an AUC of 0.70 [Q1 0.67 − Q3 0.74] and the results are considered to be significantly different according to the Wilcoxon test. All results are presented in Tab. 1.
The combination of the feature data sets did not result in an increased AUC in predicting improvement of dyspnoea. In mortality prediction, the models using the data combination showed similar AUCs using only laboratory features and all features. The median receiver operating characteristic (ROC) curves for the prediction of dyspnoea improvement (using the laboratory features) and mortality prediction (using all features) are displayed in Figs. 3 and 4, respectively.
Median receiver operating characteristic (ROC) curve from 100 Monte Carlo cross-validation iterations for the prediction of dyspnoea improvement using laboratory features. AUC area under the curve, GTB gradient tree boosting, LR logistic regression, MLP multi-layer perceptron, RFC random forest classifier, SVM support vector machine
The most relevant features for mortality prediction in GTB were determined by the importance of the features. In order of relevance, these were: NT-proBNP, BMI, CKD-EPI, creatinine, and age.
Discussion
In our population of 1,478 patients who underwent a TAVI procedure, the selected subset of ML techniques had little added prognostic value in predicting mortality and improvement of dyspnoea compared to commonly applied LR techniques. In the prediction of mortality, ML techniques achieved similar scores using all features and only the laboratory features. The increase in prognostic value and the improvement of dyspnoea prediction was rather low, even with the combination of clinical and laboratory data.
Some recent studies that applied ML have often shown positive results for prognosis prediction. In the study of Memarian et al. [21], ML methods were applied to multimodal data (clinical data, electroencephalography, magnetic resonance imaging) to predict the outcome of surgery in patients with mesial temporal lobe epilepsy, achieving a prediction accuracy of 95% using SVM-derived classifiers. Frizzell et al. [22] compared ML methods to LR in predicting 30-day readmission in patients discharged following hospitalisation for heart failure. Similar to our findings these results did not show an improvement in prediction accuracy. The prediction of six cardiovascular outcomes (including heart failure and all-cause death) was assessed by Ambale-Venkatesh et al. [23], whereby random survival forests and other ML techniques were compared to the standard cardiovascular scores. They concluded that ML improved the accuracy of cardiovascular event prediction in initially asymptomatic patients.
Our results confirm that predicting outcomes of TAVI procedures is challenging. Many factors may impact the patient’s outcome, many of which are not considered in the modelling. The inclusion of more and different kinds of features, such as different examinations, CT scans, and ECG, is currently a subject of investigation. By including different sets of features and more complex models, the predictive value may increase.
There was no implicit order in the data variables that we tried to exploit. Also, no variables were transformed into a dense representation. We included all variables that were considered relevant by clinical experts. We are aware that one-hot encoding generates data sparsity. Even though one-hot encoding can downgrade the performance of some ML methods, it is an important step for distance-based methods such as the SVM. In our study, only a small number of categorical variables (with few classes) were one-hot encoded to prevent hampering the performance of methods due to data sparsity.
The methods used in this study are generalisable to other clinical challenges in which prediction of outcomes is warranted. It is expected that the application of ML techniques in combination with clinical knowledge will become increasingly important in coming years to improve prognostics. Models with higher accuracy may improve outcome prediction after TAVI, allowing a more individual approach in clinical care.
This study suffered from a number of limitations. The dataset used in this study may be one of the largest Dutch single-centre TAVI datasets available. However, with unbalanced measures (such as a relatively small population that did not survive the 1st year), the effect of the data is reduced. Moreover, many patients were excluded because of missing data, which can be mitigated by using imputation techniques. In this study, we chose symptom reduction using the NYHA classification and 1‑year mortality as outcome measures. Other outcome measures, however, might be relevant for the TAVI population.
Conclusion
In our population of patients treated with TAVI, ML techniques were able to predict mortality using the current set of features. In predicting a reduction of dyspnoea, the traditional LR technique outperformed the others. Adding more features or increasing the dataset size may result in a situation in which ML techniques have more added value.
References
Khalil A, Faisal A, Lai KW, Ng SC, Liew YM. 2D to 3D fusion of echocardiography and cardiac CT for TAVR and TAVI image guidance. Med Biol Eng Comput. 2017;55:1317–26.
Grbic S, Mansi T, Ionasec R, et al. Image-based computational models for TAVI planning: from CT images to implant deployment. Med Image Comput Comput Interv. IEEE Trans Med Imaging. 2013;395:402.
Swee JKY, Implantation GS. (TAVI) planning from CT with ShapeForest. Med Image Comput Comput Interv—MICCAI. IEEE Trans Med Imaging. 2014;17:24.
Puri R, Iung B, Cohen DJ, Rodés-Cabau J. TAVI or no TAVI: identifying patients unlikely to benefit from transcatheter aortic valve implantation. Eur Heart J. 2016;37:2217–25.
Martin GP, Sperrin M, Ludman PF, et al. Inadequacy of existing clinical prediction models for predicting mortality after transcatheter aortic valve implantation. Am Heart J. 2017;184:97–105.
Van Mourik MS, Vendrik J, Abdelghani M, et al. Guideline-defined futility or patient-reported outcomes to assess treatment success after TAVI: what to use? Results from a prospective cohort study with long-term follow-up. Open Heart. 2018;5(2):e000879. https://doi.org/10.1136/openhrt-2018-000879.
Lebedev AV, Westman E, Van Westen GJP, et al. Random forest ensembles for detection and prediction of Alzheimer’s disease with a good between-cohort robustness. Neuroimage Clin. 2014;6:115–25.
Nishio M, Nishizawa M, Sugiyama O, et al. Computer-aided diagnosis of lung nodule using gradient tree boosting and Bayesian optimization. PLoS ONE. 2018;13:1–13.
Singal A, Mukherjee A, Elmunzer BJ, et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2016;42:407–20.
O’Brien SM, Shahian DM, et al. The Society of Thoracic Surgeons 2008 cardiac surgery risk models: part 2—isolated valve surgery. Ann Thorac Surg. 2009;88(1 Suppl):S23–S42. Jul.
Lemeshow S, Gauducheau E, et al. European system for cardiac operative risk evaluation (EuroSCORE). Eur J Cardiothorac Surg. 1999;16:9–13. Jul.
Pilgrim T, Franzone A, Stortecky S, et al. Predicting mortality after transcatheter aortic valve replacement: external validation of the transcatheter valve therapy registry model. Circ Cardiovasc Interv. 2017;10:1–9.
Ludman PF, Moat N, De Belder MA, et al. Transcatheter aortic valve implantation in the United Kingdom: temporal trends, predictors of outcome, and 6‑year follow-up: a report from the UK transcatheter aortic valve implantation (TAVI) registry, 2007 to 2012. Circulation. 2015;131:1181–90.
Lantelme P, Eltchaninoff H, Rabilloud M, et al. Development of a risk score based on aortic calcification to predict 1‑year mortality after transcatheter aortic valve replacement. Jacc Cardiovasc Imaging. 2019;12:123–32.
Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2012;12:2825–30.
Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Bishop CM. Neural networks for pattern recognition. New York: Oxford University Press; 1995.
Chen T, XGBoost GC. a scalable tree boosting system. Proc 22nd. SIGKDD Int Conf Knowl Discov Data Min. New York: ACM: ACM; 2016. pp. 785–94.
Friedman JH, Meulman JJ. Multiple additive regression trees with application in epidemiology. Stat Med. 2003;22:1365–81.
Memarian N, Kim S, Dewar S, Engel J, Staba RJ. Multimodal data and machine learning for surgery outcome prediction in complicated cases of mesial temporal lobe epilepsy. Comput Biol Med. 2015;64:67–78.
Frizzell JD, Liang L, Schulte PJ, et al. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: comparison of machine learning and other statistical approaches. Jama Cardiol. 2017;2:204–9.
Ambale-Venkatesh B, Yang X, Wu CO, et al. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res. 2017;121:1092–101.
Funding
Part of this work was funded by ITEA3 16017 project PARTNER.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
R.R. Lopes, M.S. van Mourik, E.V. Schaft, L.A. Ramos, J. Baan Jr., J. Vendrik, B.A.J.M. de Mol, M.M. Vis and H.A. Marquering declare that they have no competing interests.
Caption Electronic Supplementary Material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lopes, R.R., van Mourik, M.S., Schaft, E.V. et al. Value of machine learning in predicting TAVI outcomes. Neth Heart J 27, 443–450 (2019). https://doi.org/10.1007/s12471-019-1285-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12471-019-1285-7