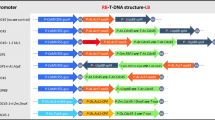
Abstract
This study was conducted to assess the suitability of the Delila gene (Del) from snapdragon for use as a visible selectable marker, under the control of the cauliflower mosaic virus (CaMV) 35S promoter, to develop a plant genetic transformation system that helps to avoid using antibiotic- or herbicide-resistance genes, such as the gene for resistance against kanamycin or PPT. Following transformation, tobacco shoots showing red coloration always contained the Del gene, which was confirmed by PCR analysis. No chimerism or ploidy variation was observed during genetic transformation. In addition, the integration ratio of the transgene to the T1 progeny was 3:1, following typical Mendelian fashion. By anthocyanin analysis, the plants containing the Del gene were shown to have 80 times higher anthocyanin content than the control plants. Thus, we conclude that strong anthocyanin accumulation, as a result of the snapdragon Del gene, can be used as a visible selectable marker for genetic transformation in the tobacco plant, replacing the use of antibiotic- or herbicide-resistance genes.
Similar content being viewed by others
References
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2394
Butelli E, Titta L, Giorgo M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C (2009) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26:1301–1308
Daley M, Knauf VC, Summerfelt KR, Turner JC (1998) Cotransformation with one Agrobacterium tumefaciens strain containing two binary plasmids as a method for producing marker-free transgenic plants. Plant Cell Rep 17:489–496
Ellul P, Garcia-Sogo B, Pineda B, Rios G, Roig, Moreno V (2003) The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum L.Mill.) is genotype and procedure dependent. Theor Appl Genet 106:231–238
Flachowsky H, Riedel M, Reim S, Hanke MV (2008) Evaluation of the uniformity and stability of T-DNA integration and gene expression in transgenic apple plants. Electron J Biotechnol 11:1–15
Geekiyanage S, Takase T, Ogura Y, Kiyosue (2007) Anthocyanin production by over-expression of grape transcription factor gene VlmybA2 in transgenic tobacco and Arabidopsis. Plant Biotechnol Rep 1:11–48
Kim CY, Ahn YO, Kim SH, Kim YH, Lee HS, Catanach AS, Jacobs JME, Conner AJ, Kwak SS (2010) The sweet potato IbMYB1 gene as a potential visible marker for sweet potato intragenic vector system. Physiol Plant 139:229–240
Kortstee AJ, Khan SA, Helderman C, Trindade LM, Wu Y, Visser RGF, Allan A, Schouten HJ, Jacobsen E (2011) Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res 20:1253–1264
Li H, Flachowsky H, Fischer TC, Hanke MV, Forkmann G, Treutter D, Schwab W, Hoffmann T, Szankowski I (2007) Maize Lc transcription factor enhances biosynthesis of anthocyanins distinct proanthocyanins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226:1243–1254
Li B, Xie C, Qiu H (2009) Production of selectable-marker-free transgenic tobacco plants using a non-selection approach: chimerism or escape, transgene inheritance, and efficiency. Plant Cell Rep 28:373–386
Lim SH, Sohn SH, Kim DH, Kim JK, Lee JY, Kim YM, Ha SH (2012) Use of an anthocyanin production phenotype as a visible selection marker system in transgenic tobacco plant. Plant Biotechnol Rep 6:203–211
Lutz KA, Svab Z, Maliga P (2006) Construction of marker-free transplastomic tobacco using the Cre-loxP site-specific recombination system. Nat Protoc 1:900–910
Maligeppagol M, Chandra GS, Navale PM, Deepa H, Rajeev PR, Asokan R, Babu KP, Barbu CSB, Rao VK, Kumar NKK (2013) Anthocyanin enrichment of tomato (Solanum lycopersicum L.) fruit by metabolic engineering. Curr Sci 105:72–80
McCormac AC, Fowler MR, Chen DF, Elliott MC (2001) Efficient co-transformation of Nicotiana tabacum by two independent TDNAs, the effect of T-DNA size and implications for genetic separation. Trans Res 10:143–155
Miki B, McHugh S (2004) Selectable marker genes in transgenic plants:applications, alternatives and biosafety. J Biotechnol 107: 193–232
Mooney M, Desnos T, Harrison K, Jones, Carpenter R, Coen E (1995) Altered regulation of tomato and tobacco pigmentation genes caused by the delila gene of Antirrhinum. Plant J 7:333–339
Naing AH, Chung JD, Park IS, Lim KB (2011) Efficient plant regeneration of the endangered medicinal orchid, Coelogyne cristata using protocorm-like bodies. Acta Physiol Plant 33:659–666
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schmulling T, Schell J (1993) Transgenic tobacco plants regenerated from leaf disks can be periclinal chimeras. Plant Mol Biol 21: 705–708
Schaart JG, Krens FA, Pelgrom KTB, Mendes O, Rouwendaal GJA (2004) Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol J 2(3):233–240
Smulders MJM, Rus-Kortekaas W, Gilissen LJW (1994) Development of polysomaty during differentiation in diploid and tetraploid tomato (Lycopersicon esculentum) plants. Plant Sci 97:53–60
Smulders MJM, Rus-Kortekaas W, Gilissen LJW (1995) Natural variation in patterns of polysomaty among individual tomato plants and their regenerated progeny. Plant Sci 106:129–139
Sreekala C, Wu J, Gu K, Wang D, Tian D, Yin Z (2005) Excision of a selectable marker in transgenic rice (Oryza sativa L.) using a chemically regulated Cre/loxP system. Plant Cell Rep 24:86–94
Vetten N, Wolters AM, Raemakers K, van der Meer I, Stege R, Heeres E, Heeres P, Visser R (2003) A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat Biotechnol 21:439–442
Ultzen T, Gielen J, Venema F, Westerbroek A, De Haan P, Tan M, Schram A, Van Grinsven M, Golbach R (1995) Resistence to tomato spotted wilt virus in transgenic tomato hybrids. Euphytica 85:159–168
Zubko E, Scutt C, Meyer P (2000) Intrachromosomal recombination between attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat Biotechnol 18:442–445
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Naing, A.H., Lim, K.B. & Kim, C.K. The usage of snapdragon Delila (Del) gene as a visible selection marker for the antibiotic-free transformation system. J. Plant Biol. 58, 110–116 (2015). https://doi.org/10.1007/s12374-014-0423-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-014-0423-4