Abstract
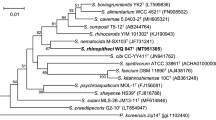
A polyphasic taxonomic approach was used to characterize three novel bacterial strains, designated as HDW12AT, HDW-15BT, and HDW15CT, isolated from the intestine of fish species Odontobutis interrupta or Siniperca scherzeri. All isolates were obligate aerobic, non-motile bacteria, and grew optimally at 30°C. Phylogenetic analysis based on 16S rRNA sequences revealed that strain HDW12AT was a member of the genus Nocardioides, and closely related to Nocardioides allogilvus CFH 30205T (98.9% sequence identities). Furthermore, strains HDW15BT and HDW15CT were members of the genus Sphingomonas, and closely related to Sphingomonas lutea JS5T and Sphingomonas sediminicola Dae 20T (97.1% and 97.9% sequence identities), respectively. Strain HDW12AT contained MK-8 (H4), and strains HDW15BT and HDW15CT contained Q-10 as the respiratory quinone. Major polar lipid components of strain HDW12AT were diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylinositol, and those of strains HDW15BT and HDW15CT were sphingoglycolipid, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and phosphatidylcholine. The G + C content of strains HDW12AT, HDW15BT, and HDW15CT were 69.7, 63.3, and 65.5%, respectively. The results of phylogenetic, phenotypic, chemotaxonomic, and genotypic analyses suggest that strain HDW12AT represents a novel species within the genus Nocardioides, and strains HDW15BT and HDW15CT represent two novel species within the genus Sphingomonas. We propose the names Nocardioides piscis for strain HDW12AT (= KACC 21336T = KCTC 49321T = JCM 33670T), Sphingomonas piscis for strain HDW15BT (= KACC 21341T = KCTC 72588T = JCM 33738T), and Sphingomonas sinipercae for strain HDW15CT (= KACC 21342T = KCTC 72589T = JCM 33739T).
Similar content being viewed by others
References
Benson, H. 1994. Microbiological Application: Laboratory manual in General Microbiology. Wan C. Publishers. Dubuque, USA.
Bousfield, G.R., Sugino, H., and Ward, D.N. 1985. Demonstration of a COOH-terminal extension on equine lutropin by means of a common acid-labile bond in equine lutropin and equine chorionic gonadotropin. J. Biol. Chem. 260, 9531–9533.
Buonaurio, R., Stravato, V.M., Kosako, Y., Fujiwara, N., Naka, T., Kobayashi, K., Cappelli, C., and Yabuuchi, E. 2002. Sphingomonas melonis sp. nov., a novel pathogen that causes brown spots on yellow Spanish melon fruits. Int. J. Syst. Evol. Microbiol. 52, 2081–2087.
Busse, H.J., Denner, E.B.M., Buczolits, S., Salkinoja-Salonen, M., Bennasar, A., and Kämpfer, P. 2003. Sphingomonas aurantiaca sp. nov., Sphingomonas aerolata sp. nov. and Sphingomonas faeni sp. nov., air- and dustborne and Antarctic, orange-pigmented, psychrotolerant bacteria, and emended description of the genus Sphingomonas. Int. J. Syst. Evol. Microbiol. 53, 1253–1260.
Chen, I.M.A., Chu, K., Palaniappan, K., Ratner, A., Huang, J., Huntemann, M., Hajek, P., Ritter, S., Varghese, N., Seshadri, R., et al. 2020. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763.
Chen, N., Jiang, J., Gao, X., Li, X., Zhang, Y., Liu, X., Yang, H., Bing, X., and Zhang, X. 2018. Histopathological analysis and the immune related gene expression profiles of mandarin fish (Siniperca chuatsi) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 83, 410–415.
Chen, H., Jogler, M., Rohde, M., Klenk, H.P., Busse, H.J., Tindall, B.J., Spröer, C., and Overmann, J. 2012. Reclassification and emended description of Caulobacter leidyi as Sphingomonas leidyi comb. nov., and emendation of the genus Sphingomonas. Int. J. Syst. Evol. Microbiol. 62, 2835–2843.
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D.R., da Costa, M.S., Rooney, A.P., Yi, H., Xu, X.W., De Meyer, S., et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466.
Collins, M.D. and Jones, D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45, 316–354.
Contreras-Moreira, B. and Vinuesa, P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701.
Dong, K., Lu, S., Yang, J., Pu, J., Lai, X.H., Jin, D., Li, J., Zhang, G., Wang, X., Liang, J., et al. 2020. Nocardioides jishulii sp. nov., isolated from faeces of Tibetan gazelle (Procapra picticaudata). Int. J. Syst. Evol. Microbiol. 70, 3665–3672.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Biol. 20, 406–416.
Fredrickson, J.K., Balkwill, D.L., Drake, G.R., Romine, M.F., Ringelberg, D.B., and White, D.C. 1995. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Appl. Environ. Microbiol. 61, 1917–1922.
Goszczynska, T. and Serfontein, J. 1998. Milk-Tween agar, a semi-selective medium for isolation and differentiation of Pseudomonas syringae pv. syringae, Pseudomonas syringae pv. phaseolicola and Xanthomonas axonopodis pv. phaseoli. J. Microbiol. Methods 32, 65–72.
Ha, S.M., Kim, C.K., Roh, J., Byun, J.H., Yang, S.J., Choi, S.B., Chun, J., and Yong, D. 2019. Application of the whole genome-based bacterial identification system, TrueBac ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann. Lab. Med. 39, 530–536.
Hiraishi, A., Ueda, Y., Ishihara, J., and Mori, T. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. Gen. Appl. Microbiol. 42, 457–469.
Hyun, D.W., Tak, E.J., Kim, P.S., and Bae, J.W. 2021. Description of Vagococcus coleopterorum sp. nov., isolated from the intestine of the diving beetle, Cybister lewisianus, and Vagococcus hydrophili sp. nov., isolated from the intestine of the dark diving beetle, Hydrophilus acuminatus, and emended description of the genus Vagococcus. J. Microbiol. 59, 132–141.
Kilic, A., Senses, Z., Kurekci, A.E., Aydogan, H., Sener, K., Kismet, E., and Basustaoglu, A.C. 2007. Nosocomial outbreak of Sphingomonas paucimobilis bacteremia in a hemato/oncology unit. Jpn. J. Infect. Dis. 60, 394–396.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lane, D. 1991. 16S/23S rRNA sequencing. In Stackbrandt, E. and Goodfellow, M. (eds.), Nucleic acid techniques in bacterial systematics, pp. 115–175. John Wiley & Sons Ltd., Hoboken, New Jersey, USA.
Lee, S., Lee, W., Chung, H.M., and Park, S. 2017. Nocardioides suum sp. nov. isolated from the air environment in an indoor pig farm. J. Microbiol. 55, 417–420.
Lee, I., Ouk Kim, Y., Park, S.C., and Chun, J. 2016. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103.
Li, H., Zhou, Y., Ling, H., Luo, L., Qi, D., and Feng, L. 2019. The effect of dietary supplementation with Clostridium butyricum on the growth performance, immunity, intestinal microbiota and disease resistance of tilapia (Oreochromis niloticus). PLoS ONE 14, e0223428.
Lin, S.Y., Wen, C.Z., Hameed, A., Liu, Y.C., Hsu, Y.H., Shen, F.T., Lai, W.A., and Young, C.C. 2015. Nocardioides echinoideorum sp. nov., isolated from sea urchins (Tripneustes gratilla). Int. J. Syst. Evol. Microbiol. 65, 1953–1958.
Liu, Y.H., Fang, B.Z., Mohamad, O.A.A., Zhang, Y.G., Jiao, J.Y., Dong, Z.Y., Xiao, M., Li, L., and Li, W.J. 2019. Nocardioides ferulae sp. nov., isolated from root of an endangered medicinal plant Ferula songorica Pall. ex Spreng. Int. J. Syst. Evol. Microbiol. 69, 1253–1258.
Liu, S.W., Xue, C.M., Li, F.N., and Sun, C.H. 2020. Nocardioides vastitatis sp. nov., isolated from Taklamakan desert soil. Int. J. Syst. Evol. Microbiol. 70, 77–82.
Matsumura, Y., Akahira-Moriya, A., and Sasaki-Mori, M. 2015. Bioremediation of bisphenol-A polluted soil by Sphingomonas bisphenolicum AO1 and the microbial community existing in the soil. Biocontrol Sci. 20, 35–42.
MIDI. 1999. Sherlock Microbial Identification System Operating Manual, version 3.0. MIDI, Inc., Newark, Delaware, USA.
Na, S.I., Kim, Y.O., Yoon, S.H., Ha, S., Baek, I., and Chun, J. 2018. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285.
Parte, A.C., Sardà Carbasse, J., Meier-Kolthoff, J.P., Reimer, L.C., and Göker, M. 2020. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612.
Pękala-Safińska, A. 2018. Contemporary threats of bacterial infections in freshwater fish. J. Vet. Res. 62, 261–267.
Picchietti, S., Miccoli, A., and Fausto, A.M. 2021. Gut immunity in European sea bass (Dicentrarchus labrax): a review. Fish Shellfish Immunol. 108, 94–108.
Piutti, S., Semon, E., Landry, D., Hartmann, A., Dousset, S., Lichtfouse, E., Topp, E., Soulas, G., and Martin-Laurent, F. 2003. Isolation and characterisation of Nocardioides sp. SP12, an atrazine-degrading bacterial strain possessing the gene trzN from bulk- and maize rhizosphere soil. FEMS Microbiol. Lett. 221, 111–117.
Prauser, H. 1976. Nocardioides, a new genus of the order Actinomycetales. Int. J. Syst. Evol. Microbiol. 26, 58–65.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, Delaware, USA.
Schleifer, K.H. and Kandler, O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477.
Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Su, S.Q., Zhang, H.Q., He, Z.Y., and Zhang, Z.X. 2005. A comparative study of the nutrients and amino acid composition of the muscle of Siniperca chuatsi and Siniperca scherzeri. Journal of Southwest Agricultural University 27, 898–901.
Teather, R.M. and Wood, P.J. 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43, 777–780.
Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Tian, J., Sun, B., Luo, Y., Zhang, Y., and Nie, P. 2009. Distribution of IgM, IgD and IgZ in mandarin fish, Siniperca chuatsi lymphoid tissues and their transcriptional changes after Flavobacterium columnare stimulation. Aquaculture 288, 14–21.
Tindall, B. 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66, 199–202.
Tittsler, R.P. and Sandholzer, L.A. 1936. The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 31, 575–580.
Tuo, L., Dong, Y.P., Habden, X., Liu, J.M., Guo, L., Liu, X.F., Chen, L., Jiang, Z.K., Liu, S.W., Zhang, Y.B., et al. 2015. Nocardioides deserti sp. nov., an actinobacterium isolated from desert soil. Int. J. Syst. Evol. Microbiol. 65, 1604–1610.
Xiao, F., Liao, L., Xu, Q., He, Z., Xiao, T., Wang, J., Huang, J., Yu, Y., Wu, B., and Yan, Q. 2021. Host-microbiota interactions and responses to grass carp reovirus infection in Ctenopharyngodon idellus. Environ. Microbiol. 23, 431–447.
Xin, H., Itoh, T., Zhou, P., Suzuki, K., Kamekura, M., and Nakase, T. 2000. Natrinema versiforme sp. nov., an extremely halophilic archaeon from Aibi salt lake, Xinjiang, China. Int. J. Syst. Evol. Microbiol. 50, 1297–1303.
Yabuuchi, E., Yano, I., Oyaizu, H., Hashimoto, Y., Ezaki, T., and Yamamoto, H. 1990. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol. Immunol. 34, 99–119.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Yoon, J. and Park, Y. 2006. The genus Nocardioides. In Dworkin, M., Falkow, S., Schleifer, K.H., and Stackebrandt, E. (eds.), The Prokaryotes, pp. 1099–1113. Springer, New York, USA.
Zhang, M.L., Li, M., Sheng, Y., Tan, F., Chen, L., Cann, I., and Du, Z.Y. 2020. Citrobacter species increase energy harvest by modulating intestinal microbiota in fish: nondominant species play important functions. mSystems 5, e00303–20.
Zhou, C., Yang, Q., and Cai, D. 1988. On the classification and distribution of the Sinipercinae fishes (Family Serranidae). Zool. Res. 9, 113–125.
Zhou, X.Y., Zhang, L., Su, X.J., Hang, P., Hu, B., and Jiang, J.D. 2019. Sphingomonas flavalba sp. nov., isolated from a procymidone-contaminated soil. Int. J. Syst. Evol. Microbiol. 69, 2936–2941.
Acknowledgements
We thank Dr. Aharon Oren (The Hebrew University of Jerusalem, Israel) and Dr. Bernard Schink (University of Konstanz, Germany) for etymological advice. This work was supported by grants from the Mid-career Researcher Program (NRF-2020R1A2C3012797) funded by the Ministry of Science and ICT, and the Basic Science Research Program (NRF-2019R1A6A3A01096031) funded by the Ministry of Education through the National Research Foundation of Korea (NRF). This work was also supported by grants from the National Institute of Biological Resources (NIBR201801106), funded by the Ministry of Environment of Korea (MOE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest The authors declare that there are no conflicts of interest.
Ethical Statement All sampling conducted in this study was approved by the Institutional Animal Care and Use Committee of Kyung Hee University (permit no. KHSASP-20-221) and complied with the guidelines of the Committee.
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hyun, DW., Jeong, YS., Lee, JY. et al. Description of Nocardioides piscis sp. nov., Sphingomonas piscis sp. nov. and Sphingomonas sinipercae sp. nov., isolated from the intestine of fish species Odontobutis interrupta (Korean spotted sleeper) and Siniperca scherzeri (leopard mandarin fish). J Microbiol. 59, 552–562 (2021). https://doi.org/10.1007/s12275-021-1036-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-021-1036-5