Abstract
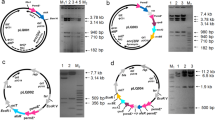
Demeclocycline (DMCTC), a tetracycline derivative antibiotic produced by Streptomyces aureofaciens, has attracted attention owing to its high bioavailability, prolonged maintenance of a therapeutic concentration, and greater efficacy against many infectious microorganisms. However, the productivity of the DMCTC-producing strains has remained low. Thus, it is necessary to identify gene-knockout or amplification targets to increase DMCTC production. Here, we demonstrated that ctcB, which encodes a Streptomyces antibiotic regulatory protein (SARP), and ctcC, which encodes a resistance gene, positively regulate the biosynthesis of DMCTC in S. aureofaciens strain DT1. In particular, overexpression of the ctcB gene in S. aureofaciens DT1 significantly enhanced DMCTC production, resulting in increased expression of ctcG, ctcN, ctcQ, ctcH, ctcV, and ctcC. The deletion of ctcB dramatically reduced the DMCTC level, implying that CtcB is an activator of DMCTC biosynthesis. Although overexpression of the ctcC, which encodes a ribosomal protection protein, enhancing DMCTC biosynthesis in S. aureofaciens DT1, the improvement was limited compared with that achieved by ctcB overexpression. This is the first study to identify the role of ctcB and ctcC in DMCTC accumulation; these genes may also be ideal candidate targets for facilitating DMCTC production by other Streptomyces strains.
Similar content being viewed by others
Reference
Choi, S. S., H. J. Nah, H. R. Pyeon, and E. S. Kim (2017) Biosynthesis, regulation, and engineering of a linear polyketide tautomycetin: a novel immunosuppressant in Streptomyces sp. CK4412. J. Ind. Microbiol. Biotechnol. 44: 555–561.
Covington, B. C., J. M. Spraggins, A. E. Ynigez-Gutierrez, Z. B. Hylton, and B. O. Bachmann (2018) Response of secondary metabolism of hypogean actinobacterial genera to chemical and biological stimuli. Appl. Environ. Microbiol. 84: e01125–18.
Liras, P. (2014) Holomycin, a dithiolopyrrolone compound produced by Streptomyces clavuligerus. Appl. Microbiol. Biotechnol. 98: 1023–1030.
Brodersen, D. E., W. M. Clemons, A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan (2000) The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 103: 1143–1154.
Duggar, B. M. (1948) Aureomycin: a product of the continuing search for new antibiotics. Ann. N.Y. Acad. Sci. 51: 177–181.
Li, H., R. Ye, G. Lin, D. Zhu, and Q. G. Mao (2016) Protein expression analysis of a high-demeclocycline producing strain of Streptomyces aureofaciens and the roles of CtcH and CtcJ in demeclocycline biosynthesis. Bioresour. Bioprocess. 3: 46.
Lein, J., L. F. Sawmiller, and L. C. Sawmiller (1959) Chlorination inhibitors affecting the biosynthesis of tetracycline. Appl. Microbiol. 7: 149–151.
Wang, I. K., L. C. Vining, J. A. Walter, and A. G. Mclnnes (1986) Use of carbon-13 in biosynthetic studies: origin of the malonyl coenzyme A incorporated into tetracycline by Streptomyces aureofaciens. J. Antibiot. 39: 1281–1287.
Dairi, T., T. Nakano, K. Aisaka, R. Katsumata, and M. Hasegawa (1995) Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci. Biotechnol. Biochem. 59: 1099–1106.
Ryan, M. J., J. A. Lotvin, N. Strathy, and S. E. Fantin (1996) Cloning of the biosynthetic pathway for chlortetracycline and tetracycline formation and cosmids useful therein. US patent 5,589,385.
Nakano, T., K. Miyake, M. Ikeda, T. Mizukami, and R. Katsumata (2000) Mechanism of the incidental production of a melanin-like pigment during 6-demethylchlortetracycline production in Streptomyces aureofaciens. Appl. Environ. Microbiol. 66: 1400–1404.
Zhu, T., X. Cheng, Y. Liu, Z. Deng, and D. You (2013) Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab. Eng. 19: 69–78.
Bibb, M. J. (2005) Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8: 208–215.
Martin, J. F. and P. Liras (2010) Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 13: 263–273.
Cho, H. S., J. C. Jo, C. H. Shin, N. Lee, J. S. Choi, B. K. Cho, J. H. Roe, C. W. Kim, H. J. Kwon, and Y. J. Yoon (2019) Improved production of clavulanic acid by reverse engineering and overexpression of the regulatory genes in an industrial Streptomyces clavuligerus strain. J. Ind. Microbiol. Biotechnol. 46: 1205–1215.
Yin, S., W. Wang, X. Wang, Y. Zhu, X. Jia, S. Li, F. Yuan, Y. Zhang, and K. Yang (2015) Identification of a cluster-situated activator of oxytetracycline biosynthesis and manipulation of its expression for improved oxytetracycline production in Streptomyces rimosus. Microb. Cell Fact. 14: 46.
Zhang, B., D. Yang, Y. Yan, G. Pan, W. Xiang, and B. Shen (2016) Overproduction of lactimidomycin by cross-overexpression of genes encoding Streptomyces antibiotic regulatory proteins. Appl. Microbiol. Biotechnol. 100: 2267–2277.
Chen, C., X. Zhao, L. Chen, Y. Jin, Z. K. Zhao, and J. W. Suh (2015) Effect of overexpression of endogenous and exogenous Streptomyces antibiotic regulatory proteins on tacrolimus (FK506) production in Streptomyces sp. KCCM11116P. RSC Adv. 5: 15765–15762.
Liu, W., Q. Zhang, J. Guo, Z. Chen, J. Li, and Y. Wen (2015) Increasing avermectin production in Streptomyces avermitilis by manipulating the expression of a novel TetR-family regulator and its target gene product. Appl. Environ. Microbiol. 81: 5157–5173.
Song, K., L. Wei, J. Liu, J. Wang, H. Qi, and J. Wen (2017) Engineering of the LysR family transcriptional regulator FkbR1 and its target gene to improve ascomycin production. Appl. Microbiol. Biotechnol. 101: 4581–4592.
Bustin, S. A., V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl, G. L. Shipley, J. Vandesompele, and C. T. Wittwer (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622.
Yang, W., L. Kong, Q. Wang, Z. Deng, and D. You (2020) Metabolic engineering of a methyltransferase for production of drug precursors demecycline and demeclocycline in Streptomyces aureofaciens. Synth. Syst. Biotechnol. 5: 121–130.
Liu, G., K. F. Chater, G. Chandra, G. Niu, and H. Tan (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77: 112–143.
Zhang, W., B. D. Ames, S. C. Tsai, and Y. Tang (2006) Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl. Environ. Microbiol. 72: 2573–2580.
Pickens, L. B., W. Kim, P. Wang, H. Zhou, K. Watanabe, S. Gomi, and Y. Tang (2009) Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. J. Am. Chem. Soc. 131: 17677–17689.
Wietzorrek, A. and M. Bibb (1997) A novel family of proteins that regulates antibiotic production in Streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25: 1181–1184.
Liu, J., T. Zhu, P. Wang, L. Kong, S. Wang, Y. Liu, C. Xie, Z. Deng, and D. You (2016) Function of Streptomyces antibiotic regulatory proteins family transcriptional regulator ctcB in the biosynthetic cluster of chlortetracycline. Acta Microbiol. Sin. 56: 1486–1495.
Wang, P., X. Gao, Y. H. Chooi, Z. Deng, and Y. Tang (2011) Genetic characterization of enzymes involved in the priming steps of oxytetracycline biosynthesis in Streptomyces rimosus. Microbiology. 157: 2401–2409.
Chen, Y., E. Wendt-Pienkowski, and B. Shen (2008) Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J. Bacteriol. 190: 5587–5596.
Tanaka, A., Y. Takano, Y. Ohnishi, and S. Horinouchi (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J. Mol. Biol. 369: 322–333.
Doyle, D., K. J. McDowall, M. J. Butler, and I. S. Hunter (1991) Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol. Microbiol. 5: 2923–2933.
Roberts, M. C. and S. Schwarz (2009) Tetracycline and chloramphenicol resistance mechanisms. pp. 183–193. In: D. L. Mayers (ed.). Antimicrobial Drug Resistance. Humana Press, New York, NY, USA.
Yin, S., X. Wang, M. Shi, F. Yuan, H. Wang, X. Jia, F. Yuan, J. Sun, T. Liu, K. Yang, Y. Zhang, K. Fan, and Z. Li (2017) Improvement of oxytetracycline production mediated via cooperation of resistance genes in Streptomyces rimosus. Sci. China Life Sci. 60: 992–999.
Wu, N., H. Huang, T. Min, and H. Hu (2017) TAR cloning and integrated overexpression of 6-demethylchlortetracycline biosynthetic gene cluster in Streptomyces aureofaciens. Acta Biochim. Biophys. Sin. 49: 1129–1134.
Wang, P., W. Kim, L. B. Pickens, X. Gao, and Y. Tang (2012) Heterologous expression and manipulation of three tetracycline biosynthetic pathways. Angew. Chem. Int. Ed. Eng. 51: 11136–11140.
Malla, S., N. P. Niraula, K. Liou, and J. K. Sohng (2010) Self-resistance mechanism in Streptomyces peucetius: Overexpression of drrA, drrB and drrC for doxorubicin enhancement. Microbiol. Res. 165: 259–267.
Chu, X., Z. Zhen, Z. Tang, Y. Zhuang, J. Chu, S. Zhang, and M. Guo (2012) Introduction of extra copy of oxytetracycline resistance gene otrB enhances the biosynthesis of oxytetracycline in Streptomyces rimosus. J. Bioprocess. Biotechniq. 2: 117.
Acknowledgments
This work was financially supported by Topfond Pharmaceutical Co., Ltd. Henan, China. We thank Professor Liujing Wei for her assistance in this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest.
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
12257_2020_284_MOESM1_ESM.pdf
Increasing Demeclocycline Production in Streptomyces aureofaciens by Manipulating the Expression of a Novel SARP Family Regulator and Its Genes
Rights and permissions
About this article
Cite this article
Tan, YY., Zhu, GY., Ye, RF. et al. Increasing Demeclocycline Production in Streptomyces aureofaciens by Manipulating the Expression of a Novel SARP Family Regulator and Its Genes. Biotechnol Bioproc E 26, 887–897 (2021). https://doi.org/10.1007/s12257-020-0284-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-020-0284-2