Abstract
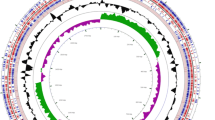
Pediococcus acidilactici is a probiotic bacterium that is industrially utilized in the food industry and antibiotics development. Here, we determine the complete nucleotide sequence of the genome of Pediococcus acidilactici ATCC 8042. The genome was sequenced by the PacBio RSII to generate a single contig consisting of circular chromosome sequence. Illumina MiniSeq sequencing platform and Sanger sequencing method were additionally utilized to correct errors resulting from the long-read sequencing platform. The sequence consists of 2,009,598 bp with a G + C content of 42.1% and contains 1,865 protein-coding sequences. Based on the sequence information, we could confirm and predict the presence of four peptidoglycan hydrolases by HyPe software. This work, therefore, provides the complete genomic information of P. acidilactici ATCC 8042 with a profitable potential of genome-scale comprehension of anti-pathogenic activity, which can be applied in nutraceutical and pharmaceutical biotechnology field.
Similar content being viewed by others
References
Charchoghlyan, H., J. Bae, H. Kwon, and K. Myunghee (2017) Rheological properties and volatile composition of fermented milk prepared by exopolysaccharide-producing Lactobacillus acidophilus n.v. Er2 317/402 strain narine. Biotechnology and Bioprocess Engineering. 22: 327–338.
Mikulski, D., J. Jankowski, J. Naczmanski, M. Mikulska, and V. Demey (2012) Effects of dietary probiotic (Pediococcus acidilactici) supplementation on performance, nutrient digestibility, egg traits, egg yolk cholesterol, and fatty acid profile in laying hens. Poult Sci. 91: 2691–2700.
Anastasiadou, S., M. Papagianni, G. Filiousis, I. Ambrosiadis, and P. Koidis (2008) Pediocin SA-1, an antimicrobial peptide from Pediococcus acidilactici NRRL B5627: production conditions, purification and characterization. Bioresour Technol. 99: 5384–5390.
Klaenhammer, T. R. (1993) Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 12: 39–85.
Ferguson, R. M., D. L. Merrifield, G. M. Harper, M. D. Rawling, S. Mustafa, S. Picchietti, et al. (2010) The effect of Pediococcus acidilactici on the gut microbiota and immune status of on-growing red tilapia (Oreochromis niloticus). J. Appl. Microbiol. 109: 851–862.
Papagianni, M. and S. Anastasiadou (2009) Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microb. Cell Fact. 8: 3.
Galvez, A., H. Abriouel, R. L. Lopez, and N. Ben Omar (2007) Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120: 51–70.
Mora, D., M. G. Fortina, C. Parini, D. Daffonchio, and P. L. Manachini (2000) Genomic subpopulations within the species Pediococcus acidilactici detected by multilocus typing analysis: relationships between pediocin AcH/PA-1 producing and non-producing strains. Microbiology. 146 (Pt 8): 2027–2038.
Garcia-Cano, I., L. Velasco-Perez, R. Rodriguez-Sanoja, S. Sanchez, G. Mendoza-Hernandez, A. Llorente-Bousquets, et al. (2011) Detection, cellular localization and antibacterial activity of two lytic enzymes of Pediococcus acidilactici ATCC 8042. J. Appl. Microbiol. 111: 607–615.
John Ndayishimiye, D. J. L. and B. S. Chun (2017) Impact of extraction conditions on bergapten content and antimicrobial activity of oils obtained by a co-extraction of citrus by-products using supercritical carbon dioxide. Biotechnology and Bioprocess Engineering. 22: 586–596.
Garcia-Cano, I., M. Campos-Gomez, M. Contreras-Cruz, C. E. Serrano-Maldonado, A. Gonzalez-Canto, C. Pena-Montes, et al. (2015) Expression, purification, and characterization of a bifunctional 99-kDa peptidoglycan hydrolase from Pediococcus acidilactici ATCC 8042. Appl Microbiol Biotechnol. 99: 8563–8573.
Optimization of technical conditions for the transformation of Pediococcus acidilactici P60 by electroporation.
Deatherage, D. E. and J. E. Barrick (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151: 165–188.
Aziz, R. K., D. Bartels, A. A. Best, M. DeJongh, T. Disz, R. A. Edwards, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 9: 75.
Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30: 2068–2069.
Huerta-Cepas, J., D. Szklarczyk, D. Heller, A. Hernandez-Plaza, S. K. Forslund, H. Cook, et al. (2018) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res.
Sharma, A. K., S. Kumar, K. Harish, D. B. Dhakan, and V. K. Sharma (2016) Prediction of peptidoglycan hydrolases- a new class of antibacterial proteins. BMC Genomics. 17: 411.
Zhu, W., A. Lomsadze, and M. Borodovsky (2010) Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38: e132.
Si, J.-B., E.-J. Jang, D. Charalampopoulos, Y.-J. Wee (2018) Purification and characterization of microbial protease produced extracellularly from Bacillus subtilis FBL-1. Biotechnology and Bioprocess Engineering. 23: 176–182.
Acknowledgements
This work was supported by the Bio & Medical Technology Development Program (NRF-2018M3A9H3020459) of the National Research Foundation (NRF) funded by Ministry of Science and ICT (MSIT) and the Next-Generation BioGreen 21 Program (PJ01323601) funded by Rural Development Administration and Creative-Pioneering Researchers Program through Seoul National University (SNU).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cho, S.W., Yang, J., Park, S. et al. Complete Genome Sequence of Lactic Acid Bacterium Pediococcus acidilactici Strain ATCC 8042, an Autolytic Anti-bacterial Peptidoglycan Hydrolase Producer. Biotechnol Bioproc E 24, 483–487 (2019). https://doi.org/10.1007/s12257-019-0037-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-019-0037-2