Abstract
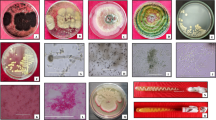
It is generally accepted that dead tree decomposition is performed mainly by delignifying basidiomycetes. While ascomycetes have been reported to inhabit dead tree bark, their contribution to dead tree decomposition is still unclear. Here, we isolated five bark-inhabiting ascomycetes possessing cellulolytic activity from dead beech tree and assessed their polysaccharolytic activities. When cultivated in a medium containing filter paper as a sole carbon source, three strains degraded >40 % of the filter paper in a 4-week cultivation and the others degraded 15–30 % of the paper. The degraders possessed amylolytic, pectinolytic, and mannanolytic activities as well as cellulolytic activity, implying that they play an important role in dead tree decomposition after delignification by basidiomycetes. Phylogenetic analysis based on large subunit ribosomal DNA (lsu-DNA) sequences implied that the isolates belonged to Penicillium or Amorphotheca.
Similar content being viewed by others
References
Abliz P., Fukushima K., Takizawa K., Nishimura K.: Identification of pathogenic dematiaceous fungi and related taxa based on large subunit ribosomal DNA D1/D2 domain sequence analysis. FEMS Immunol.Med.Microbiol.40, 41–49 (2004).
Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.: Basic local alignment search tool. J.Mol.Biol.215, 403–410 (1990).
Bååth E., Anderson T.H.: Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol.Biochem.35, 955–963 (2003).
Baldrian P.: Wood-inhabiting ligninolytic basidiomycetes in soils: ecology and constraints for applicability in bioremediation. Fungal Ecol.1, 4–12 (2008).
Baldrian P., Valašková V.: Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol.Rev.32, 501–521 (2008).
Blagodatskaya E.V., Anderson T.H.: Interactive effects of pH and substrate quality on the fungal-to-bacterial ratio and qCO2 of microbial communities in forest soils. Soil Biol.Biochem.30, 1269–1274 (1998).
Bunyard B.A., Nicholson M.S., Royse D.J.: Phylogenetic resolution of Morchella, Verpa, and Disciotis based on restriction enzyme analysis of the 28S ribosomal RNA gene. Exp.Mycol.19, 223–233 (1995).
Chen J.C., Liu Z.S.: Soil characteristics and clay mineralogy of two subalpine forest spodosols with clay accumulation in Taiwan. Soil Sci.169, 66–80 (2004).
Cofone L. Jr., Walker J.D., Cooney J.J.: Utilization of hydrocarbons by Cladosporium resinae. J.Gen.Microbiol.76, 243–246 (1973).
Deshpande V., Rao M., Keskar S., Mishra C.: Occurrence of a procellulase in the culture filtrates of Penicillium janthinellum. Enzyme Microb.Technol.6, 371–374 (1984).
Felsenstein J.: Confidence limits on phylogenies: an approach using the bootstrap. Evolution39, 783–791 (1985).
Ghose T.K.: Measurement of cellulose activities. Pure Appl.Chem.59, 257–268 (1987)
Hernández-Luna C.E., Gutiérrez-Soto G., Salcedo-martínez S.M.: Screening for decolorizing basidiomycetes in Mexico. World J.Microbiol.Biotechnol.24, 465–473 (2008).
Hinrikson H.P., Hurst S.F., Lott T.J., Warnock D.W., Morrison C.J.: Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. J.Clin.Microbiol.43, 2092–2103 (2005).
Krogh K.B.R., Mørkeberg A., Jørgensen H., Frisvad J.C., Olsson L.: Screening genus Penicillium for producers of cellulolytic and xylanolytic enzymes. Appl.Biochem.Biotechnol.113–116, 389–401 (2005).
Kubátová A.: Neglected Penicillium spp. associated with declining trees, pp. 299–308 in Integration of Modern Taxonomic methods for Penicillium and Aspergillus Classification. Taylor & Francis, London 2000.
Kurtzman C.P., Robnett C.J.: Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek73, 331–371 (1998).
Leonowicz A., Matuszewska A., Luterek J., Ziegenhagen D., Wojtas-Wasilewska M., Cho N.S., Hofrichter M.: Biodegradation of lignin by white rot fungi. Fungal Genet.Biol.27, 175–185 (1999).
Liers C., Ullrich R., Steffen K.T., Hatakka A., Hofrichter M.: Mineralization of 14C-labelled synthetic lignin and extracellular enzyme activities of the wood-colonizing ascomycetes Xylaria hypoxylon and Xylaria polymorpha. Appl.Microbiol.Biotechnol.69, 573–579 (2005).
Lopez M.J., Vargas-García M.C., Suarez-Estrellá F., Nichols N.N., Dien B.S., Moreno J.: Lignocellulose-degrading enzymes produced by the ascomycete Coniochaeta ligniaria and related species: application for a lignocellulosic substrate treatment. Enzyme Microb.Technol.40, 794–800 (2007).
Madhu G.L.S., Prabhu K.A.: Studies on dextranase from Penicillium aculeatum. Enzyme Microb.Technol.6, 217–220 (1984).
Morris D.L.: Quantitative determination of carbohydrates with Dreywoods anthrone reagent. Science107, 254–255 (1948).
O’brien H.E., Parrent J.L., Jackson J.A., Moncalvo J.M., Vilgalys R.: Fungal community analysis by large-scale sequencing of environmental samples. Appl.Environ.Microbiol.71, 5544–5550 (2005).
Osono T., Takeda H.: Comparison of litter decomposing ability among diverse fungi in a cool temperate deciduous forest in Japan. Mycologia94, 421–427 (2002).
Osono T., Takeda H.: Fungal decomposition of Abies needle and Betula leaf litter. Mycologia98, 172–179 (2006).
Pandedy S., Selvakumar P., Soccol C.R., Nigam P.: Solid state fermentation for the production of industrial enzymes. Current Sci.77, 143–162 (1999).
Rigas F., Marchant R., Dritsa V., Kapsanaki-Gotsi E., Gonou-Zagou Z., Avramides E.J.: Screening of wood rotting fungi potentially useful for degradation of organic pollutants. Water Air Soil Poll.3, 201–210 (2003).
Saha B.C., Iten L.B., Cotta M.A., Wu Y.V.: Dilute acid pretreatment, enzymatic saccharification, and fermentation of rice hulls to ethanol. Biotechnol.Progr.21, 816–822 (2005).
Saitou N., Nei M.: The neighbor-joining method: a new method for reconstructing phylogenic trees. Mol.Biol.Evol.4, 406–425 (1987).
Satomi M., Kimura B., Mizoi M., Satou T., Fujii T.: Tetragenococcus muriaticus sp.nov., a new moderately halophilic lactic acid bacterium isolated from fermented squid liver sauce. Internat.J.Syst.Bacteriol.47, 832–836 (1997).
Sheridan J.E.: Monitoring for the kerosene fungus Amorphotheca resinae. Revista Microbiol.5, 67–71 (1974).
Šnajdr J., Baldrian P.: Temperature affects the production, activity, and stability of lignolytic enzymes in Pleurotus ostreatus and Trametes versicolor. Folia Microbiol.5, 498–502 (2007).
The J.S., Lee K.H.: Utilization of n-alkanes by Cladosporium resinae. Appl.Environ.Microbiol.25, 454–457 (1973).
Thompson J.D., Higgins D.G., Gibson T.J.: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl.Acids Res.22, 4673–4680 (1994).
Timell T.E.: Recent progress in the chemistry of wood hemicelluloses. Wood Sci.Technol.1, 45–70 (1967).
Tomšovský M., Popelářová P., Baldrian P.: Production and regulation of lignocellulose-degrading enzymes of Poria-like woodinhabiting basidiomycetes. Folia Microbiol.1, 74–80 (2009).
Vidal S., Salmon J., Williams P., Pellerin P.: Penicillium daleae, a soil fungus able to degrade rhamnogalacturonan II, a complex pectic polysaccharide. Enzyme Microb.Technol.24, 283–290 (1999).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fujii, K., Sugimura, T. & Nakatake, K. Ascomycetes with cellulolytic, amylolytic, pectinolytic, and mannanolytic activities inhabiting dead beech (Fagus crenata) trees. Folia Microbiol 55, 29–34 (2010). https://doi.org/10.1007/s12223-010-0005-x
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-010-0005-x