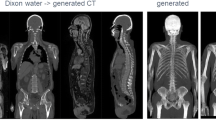
Abstract
Artificial intelligence (AI) has been applied to various medical imaging tasks, such as computer-aided diagnosis. Specifically, deep learning techniques such as convolutional neural network (CNN) and generative adversarial network (GAN) have been extensively used for medical image generation. Image generation with deep learning has been investigated in studies using positron emission tomography (PET). This article reviews studies that applied deep learning techniques for image generation on PET. We categorized the studies for PET image generation with deep learning into three themes as follows: (1) recovering full PET data from noisy data by denoising with deep learning, (2) PET image reconstruction and attenuation correction with deep learning and (3) PET image translation and synthesis with deep learning. We introduce recent studies based on these three categories. Finally, we mention the limitations of applying deep learning techniques to PET image generation and future prospects for PET image generation.
Similar content being viewed by others
References
Fukushima K, Miyake S. Neocognitron: a self-organizing neural network model for a mechanism of visual pattern recognition. Competition and cooperation in neural nets. Lecture notes in biomathematics. Springer; 1982. p. 267–85.
Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323(6088):533–6. https://doi.org/10.1038/323533a0.
Hinton GE, Sejnowski TJ. Learning and relearning in Boltzmann machines Parallel distributed processing: explorations in the microstructure of cognition. MIT Press; 1986.
Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press; 2016.
Doi K. Computer-aided diagnosis in medical imaging historical review, current status and future potential. Comput Med Imaging Gr. 2007;31(4–5):198–211. https://doi.org/10.1016/j.compmedimag.2007.02.002.
Fujita H. AI-based computer-aided diagnosis (AI-CAD): the latest review to read first. Radiol Phys Technol. 2020;13(1):6–19. https://doi.org/10.1007/s12194-019-00552-4.
Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27(9):1533–9. https://doi.org/10.1038/sj.jcbfm.9600493.
Hsu DFC, Ilan E, Peterson WT, Uribe J, Lubberink M, Levin CS. Studies of a next-generation silicon-photomultiplier-based time-of-flight PET/CT system. J Nucl Med. 2017;58(9):1511–8. https://doi.org/10.2967/jnumed.117.189514.
van Sluis J, de Jong J, Schaar J, Noordzij W, van Snick P, Dierckx R, et al. Performance characteristics of the digital biograph Vision PET/CT system. J Nucl Med. 2019;60(7):1031–6. https://doi.org/10.2967/jnumed.118.215418.
Teoh EJ, McGowan DR, Macpherson RE, Bradley KM, Gleeson FV. Phantom and clinical evaluation of the Bayesian Penalized likelihood reconstruction algorithm Q.Clear on an LYSO PET/CT system. J Nucl Med. 2015;56(9):1447–52. https://doi.org/10.2967/jnumed.115.159301.
Lee YS, Kim JS, Kim KM, Kang JH, Lim SM, Kim HJ. Performance measurement of PSF modeling reconstruction (True X) on Siemens Biograph TruePoint TrueV PET/CT. Ann Nucl Med. 2014;28(4):340–8. https://doi.org/10.1007/s12149-014-0815-z.
Wang T, Lei Y, Fu Y, Wynne JF, Curran WJ, Liu T, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys. 2021;22(1):11–36. https://doi.org/10.1002/acm2.13121.
Tian C, Fei L, Zheng W, Xu Y, Zuo W, Lin CW. Deep learning on image denoising: an overview. Neural Netw. 2020;131:251–75. https://doi.org/10.1016/j.neunet.2020.07.025.
Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys. 2017;44(10):e360–75. https://doi.org/10.1002/mp.12344.
Shan H, Padole A, Homayounieh F, Kruger U, Khera RD, Nitiwarangkul C, et al. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat Mach Intell. 2019;1(6):269–76. https://doi.org/10.1038/s42256-019-0057-9.
Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans Med Imaging. 2017;36(12):2536–45. https://doi.org/10.1109/TMI.2017.2708987.
Yi X, Babyn P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J Digit Imaging. 2018;31(5):655–69. https://doi.org/10.1007/s10278-018-0056-0.
Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017;267:406–16. https://doi.org/10.1016/j.neucom.2017.06.048.
Ronneberger O, Fischer P, Brox T. U-Net convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015. Springer International Publishing; 2015. p. 234–41.
Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, et al. Ultra-low-dose (18)F-Florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290(3):649–56. https://doi.org/10.1148/radiol.2018180940.
Schaefferkoetter J, Yan J, Ortega C, Sertic A, Lechtman E, Eshet Y, et al. Convolutional neural networks for improving image quality with noisy PET data. EJNMMI Res. 2020;10(1):105. https://doi.org/10.1186/s13550-020-00695-1.
Ladefoged CN, Hasbak P, Hornnes C, Hojgaard L, Andersen FL. Low-dose PET image noise reduction using deep learning: application to cardiac viability FDG imaging in patients with ischemic heart disease. Phys Med Biol. 2021;66(5): 054003. https://doi.org/10.1088/1361-6560/abe225.
Spuhler K, Serrano-Sosa M, Cattell R, DeLorenzo C, Huang C. Full-count PET recovery from low-count image using a dilated convolutional neural network. Med Phys. 2020;47(10):4928–38. https://doi.org/10.1002/mp.14402.
da Costa-Luis CO, Reader AJ. Micro-networks for robust MR-guided low count PET imaging. IEEE Trans Radiat Plasma Med Sci. 2021;5(2):202–12. https://doi.org/10.1109/TRPMS.2020.2986414.
Wang YJ, Baratto L, Hawk KE, Theruvath AJ, Pribnow A, Thakor AS, et al. Artificial intelligence enables whole-body positron emission tomography scans with minimal radiation exposure. Eur J Nucl Med Mol Imaging. 2021;48(9):2771–81. https://doi.org/10.1007/s00259-021-05197-3.
Chen KT, Schurer M, Ouyang J, Koran MEI, Davidzon G, Mormino E, et al. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur J Nucl Med Mol Imaging. 2020;47(13):2998–3007. https://doi.org/10.1007/s00259-020-04897-6.
Liu H, Wu J, Lu W, Onofrey JA, Liu YH, Liu C. Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose PET. Phys Med Biol. 2020;65(18): 185006. https://doi.org/10.1088/1361-6560/abae08.
Kim K, Wu D, Gong K, Dutta J, Kim JH, Son YD, et al. Penalized PET reconstruction using deep learning prior and local linear fitting. IEEE Trans Med Imaging. 2018;37(6):1478–87. https://doi.org/10.1109/TMI.2018.2832613.
Lv Y, Xi C. PET image reconstruction with deep progressive learning. Phys Med Biol. 2021;66(10): 105016. https://doi.org/10.1088/1361-6560/abfb17.
Wang X, Zhou L, Wang Y, Jiang H, Ye H. Improved low-dose positron emission tomography image reconstruction using deep learned prior. Phys Med Biol. 2021;66(11): 115001. https://doi.org/10.1088/1361-6560/abfa36.
Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–62. https://doi.org/10.1016/j.neuroimage.2018.03.045.
Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. 2019;64(16): 165019. https://doi.org/10.1088/1361-6560/ab3242.
Wang Y, Zhou L, Wang L, Yu B, Zu C, Lalush DS, et al. Locality adaptive multi-modality GANs for high-quality PET image synthesis. Medical image computing and computer assisted intervention–MICCAI 201. Springer International Publishing; 2018. p. 329–37.
Ouyang J, Chen KT, Gong E, Pauly J, Zaharchuk G. Ultra-low-dose PET reconstruction using generative adversarial network with feature matching and task-specific perceptual loss. Med Phys. 2019;46(8):3555–64. https://doi.org/10.1002/mp.13626.
Zhu J-Y, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. 2017 IEEE International Conference on Computer Vision (ICCV). Venice, Italy: IEEE; 2017. p. 2242–51.
Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran WJ, et al. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Phys Med Biol. 2019;64(21): 215017. https://doi.org/10.1088/1361-6560/ab4891.
Sanaat A, Shiri I, Arabi H, Mainta I, Nkoulou R, Zaidi H. Deep learning-assisted ultra-fast/low-dose whole-body PET/CT imaging. Eur J Nucl Med Mol Imaging. 2021;48(8):2405–15. https://doi.org/10.1007/s00259-020-05167-1.
Zhao K, Zhou L, Gao S, Wang X, Wang Y, Zhao X, et al. Study of low-dose PET image recovery using supervised learning with CycleGAN. PLoS ONE. 2020;15(9): e0238455. https://doi.org/10.1371/journal.pone.0238455.
Zhou L, Schaefferkoetter JD, Tham IWK, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Med Image Anal. 2020;65: 101770. https://doi.org/10.1016/j.media.2020.101770.
Ulyanov D, Vedaldi A, Lempitsky V. Deep Image Prior [https://arxiv.org/abs/quant-ph/1711.10925]. 2017.
Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019;46(13):2780–9. https://doi.org/10.1007/s00259-019-04468-4.
Gong K, Catana C, Qi J, Li Q. PET image reconstruction using deep image prior. IEEE Trans Med Imaging. 2019;38(7):1655–65. https://doi.org/10.1109/TMI.2018.2888491.
Hashimoto F, Ohba H, Ote K, Teramoto A, Tsukada H. Dynamic PET image denoising using deep convolutional neural networks without prior training datasets. IEEE Access. 2019;7:96594–603. https://doi.org/10.1109/access.2019.2929230.
Hashimoto F, Ohba H, Ote K, Kakimoto A, Tsukada H, Ouchi Y. 4D deep image prior: dynamic PET image denoising using an unsupervised four-dimensional branch convolutional neural network. Phys Med Biol. 2021;66(1): 015006. https://doi.org/10.1088/1361-6560/abcd1a.
Wang T, Lei Y, Fu Y, Curran WJ, Liu T, Nye JA, et al. Machine learning in quantitative PET: a review of attenuation correction and low-count image reconstruction methods. Phys Med. 2020;76:294–306. https://doi.org/10.1016/j.ejmp.2020.07.028.
Shiyam Sundar LK, Muzik O, Buvat I, Bidaut L, Beyer T. Potentials and caveats of AI in hybrid imaging. Methods. 2021;188:4–19. https://doi.org/10.1016/j.ymeth.2020.10.004.
Arabi H, AkhavanAllaf A, Sanaat A, Shiri I, Zaidi H. The promise of artificial intelligence and deep learning in PET and SPECT imaging. Phys Med. 2021;83:122–37. https://doi.org/10.1016/j.ejmp.2021.03.008.
Zaharchuk G, Davidzon G. Artificial Intelligence for Optimization and Interpretation of PET/CT and PET/MR Images. Semin Nucl Med. 2021;51(2):134–42. https://doi.org/10.1053/j.semnuclmed.2020.10.001.
Lee JS. A review of deep-learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Rad Plasma Med Sci. 2021;5(2):160–84. https://doi.org/10.1109/trpms.2020.3009269.
Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature. 2018;555(7697):487–92. https://doi.org/10.1038/nature25988.
Haggstrom I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder-decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–62. https://doi.org/10.1016/j.media.2019.03.013.
Wang B, Liu H. FBP-Net for direct reconstruction of dynamic PET images. Phys Med Biol. 2020. https://doi.org/10.1088/1361-6560/abc09d.
Whiteley W, Panin V, Zhou C, Cabello J, Bharkhada D, Gregor J. FastPET: near real-time reconstruction of PET histo-image data using a neural network. IEEE Trans Rad Plasma Med Sci. 2021;5(1):65–77. https://doi.org/10.1109/trpms.2020.3028364.
Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. J Med Imaging (Bellingham). 2020;7(3): 032503. https://doi.org/10.1117/1.JMI.7.3.032503.
Hashimoto F, Ito M, Ote K, Isobe T, Okada H, Ouchi Y. Deep learning-based attenuation correction for brain PET with various radiotracers. Ann Nucl Med. 2021;35(6):691–701. https://doi.org/10.1007/s12149-021-01611-w.
Shiri I, Arabi H, Geramifar P, Hajianfar G, Ghafarian P, Rahmim A, et al. Deep-JASC: joint attenuation and scatter correction in whole-body (18)F-FDG PET using a deep residual network. Eur J Nucl Med Mol Imaging. 2020;47(11):2533–48. https://doi.org/10.1007/s00259-020-04852-5.
Arabi H, Bortolin K, Ginovart N, Garibotto V, Zaidi H. Deep learning-guided joint attenuation and scatter correction in multitracer neuroimaging studies. Hum Brain Mapp. 2020;41(13):3667–79. https://doi.org/10.1002/hbm.25039.
Yang J, Park D, Gullberg GT, Seo Y. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain (18)F-FDG PET. Phys Med Biol. 2019;64(7): 075019. https://doi.org/10.1088/1361-6560/ab0606.
Yang J, Sohn JH, Behr SC, Gullberg GT, Seo Y. CT-less direct correction of attenuation and scatter in the image space using deep learning for whole-body FDG PET: potential benefits and pitfalls. Radiol Artif Intell. 2021;3(2): e200137. https://doi.org/10.1148/ryai.2020200137.
Choi H, Lee DS. Alzheimer’s disease neuroimaging I. Generation of structural MR images from Amyloid PET: application to MR-less quantification. J Nucl Med. 2018;59(7):1111–7. https://doi.org/10.2967/jnumed.117.199414.
Kang SK, Seo S, Shin SA, Byun MS, Lee DY, Kim YK, et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 2018;39(9):3769–78. https://doi.org/10.1002/hbm.24210.
Kimura Y, Watanabe A, Yamada T, Watanabe S, Nagaoka T, Nemoto M, et al. AI approach of cycle-consistent generative adversarial networks to synthesize PET images to train computer-aided diagnosis algorithm for dementia. Ann Nucl Med. 2020;34(7):512–5. https://doi.org/10.1007/s12149-020-01468-5.
Kang SK, Choi H, Lee JS. Alzheimer’s disease neuroimaging initiative translating amyloid PET of different radiotracers by a deep generative model for interchangeability. Neuroimage. 2021;232:117890. https://doi.org/10.1016/j.neuroimage.2021.117890.
Guo J, Gong E, Fan AP, Goubran M, Khalighi MM, Zaharchuk G. Predicting (15)O-Water PET cerebral blood flow maps from multi-contrast MRI using a deep convolutional neural network with evaluation of training cohort bias. J Cereb Blood Flow Metab. 2020;40(11):2240–53. https://doi.org/10.1177/0271678X19888123.
Chen DYT, Ishii Y, Fan AP, Guo J, Zhao MY, Steinberg GK, et al. Predicting PET Cerebrovascular Reserve with Deep Learning by Using Baseline MRI: a pilot investigation of a drug-free brain stress test. Radiology. 2020;296(3):627–37. https://doi.org/10.1148/radiol.2020192793.
Matsubara K, Ibaraki M, Shinohara Y, Takahashi N, Toyoshima H, Kinoshita T. Prediction of an oxygen extraction fraction map by convolutional neural network: validation of input data among MR and PET images. Int J Comput Assist Radiol Surg. 2021;16(11):1865–74. https://doi.org/10.1007/s11548-021-02356-7.
Ben-Cohen A, Klang E, Raskin SP, Soffer S, Ben-Haim S, Konen E, et al. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng Appl Artif Intell. 2019;78:186–94. https://doi.org/10.1016/j.engappai.2018.11.013.
Sanaat A, Mirsadeghi E, Razeghi B, Ginovart N, Zaidi H. Fast dynamic brain PET imaging using stochastic variational prediction for recurrent frame generation. Med Phys. 2021;48(9):5059–71. https://doi.org/10.1002/mp.15063.
Wang R, Liu H, Toyonaga T, Shi L, Wu J, Onofrey JA, et al. Generation of synthetic PET images of synaptic density and amyloid from (18) F-FDG images using deep learning. Med Phys. 2021;48(9):5115–29. https://doi.org/10.1002/mp.15073.
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. An unsupervised learning model for deformable medical image registration. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Salt Lake City, UT, USA: IEEE; 2018. p. 9252–60.
Hering A, Kuckertz S, Heldmann S, Heinrich MP. Memory-efficient 2.5D convolutional transformer networks for multi-modal deformable registration with weak label supervision applied to whole-heart CT and MRI scans. Int J Comput Assist Radiol Surg. 2019;14(11):1901–12. https://doi.org/10.1007/s11548-019-02068-z.
Yang G, Wang C, Yang J, Chen Y, Tang L, Shao P, et al. Weakly-supervised convolutional neural networks of renal tumor segmentation in abdominal CTA images. BMC Med Imaging. 2020;20(1):37. https://doi.org/10.1186/s12880-020-00435-w.
Kallenberg M, Petersen K, Nielsen M, Ng AY, Pengfei D, Igel C, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016;35(5):1322–31. https://doi.org/10.1109/TMI.2016.2532122.
Spitzer H, Kiwitz K, Amunts K, Harmeling S, Dickscheid T. Improving Cytoarchitectonic Segmentation of Human Brain Areas with Self-supervised Siamese Networks [https://arxiv.org/abs/quant-ph/1806.05104]. 2018.
Caron M, Bojanowski P, Joulin A, Douze M. Deep Clustering for Unsupervised Learning of Visual Features [https://arxiv.org/abs/quant-ph/1807.05520]. 2018.
Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA. Context Encoders: Feature Learning by Inpainting [https://arxiv.org/abs/quant-ph/1604.07379]. 2016.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention Is All You Need [https://arxiv.org/abs/quant-ph/1706.03762]. 2017.
Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding [https://arxiv.org/abs/quant-ph/1810.04805]. 2018.
Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov R, Le QV. XLNet: Generalized Autoregressive Pretraining for Language Understanding [https://arxiv.org/abs/quant-ph/1906.08237]. 2019.
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale [https://arxiv.org/abs/quant-ph/2010.11929]. 2020.
Jiang Y, Chang S, Wang Z. TransGAN: Two Pure Transformers Can Make One Strong GAN, and That Can Scale Up [https://arxiv.org/abs/quant-ph/2102.07074]. 2021.
Watanabe S, Ueno T, Kimura Y, Mishina M, Sugimoto N. Generative image transformer (GIT): unsupervised continuous image generative and transformable model for [(123)I]FP-CIT SPECT images. Ann Nucl Med. 2021;35(11):1203–13. https://doi.org/10.1007/s12149-021-01661-0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matsubara, K., Ibaraki, M., Nemoto, M. et al. A review on AI in PET imaging. Ann Nucl Med 36, 133–143 (2022). https://doi.org/10.1007/s12149-021-01710-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12149-021-01710-8