Abstract
Stochastic ecological network occupancy (SENO) models predict the probability that species will occur in a sample of an ecological network. In this review, we introduce SENO models as a means to fill a gap in the theoretical toolkit of ecologists. As input, SENO models use a topological interaction network and rates of colonization and extinction (including consumer effects) for each species. A SENO model then simulates the ecological network over time, resulting in a series of sub-networks that can be used to identify commonly encountered community modules. The proportion of time a species is present in a patch gives its expected probability of occurrence, whose sum across species gives expected species richness. To illustrate their utility, we provide simple examples of how SENO models can be used to investigate how topological complexity, species interactions, species traits, and spatial scale affect communities in space and time. They can categorize species as biodiversity facilitators, contributors, or inhibitors, making this approach promising for ecosystem-based management of invasive, threatened, or exploited species.
Similar content being viewed by others
Introduction
Artist Paul Klee felt “Nature is garrulous to the point of confusion”. This confusion inspires biologists as well as artists, and ecologists have devoted an increasing amount of research to complex networks of interacting species and the implications of network topology and dynamics for ecological function and stability (Dunne 2006; Montoya et al. 2006; Bascompte 2009). Ecological networks characterize numerous direct and indirect effects that are difficult if not impossible to predict solely from studies of predator–prey dynamics or competition among guild members (O'Gorman and Emmerson 2009). The challenge of analyzing networks is that the dimension of the problem increases rapidly with species richness. In this paper, we describe a new approach, stochastic ecological network occupancy (SENO) models, for modeling ecological networks. The approach integrates species extinctions and colonizations into network modeling. Tracking occupancy in networks over time provides the expected probability that each species will occur in a sample (Fig. 1).
Illustration of a SENO model from t to t + 20. Extinctions (solidus) are stochastic with expected rates. In comparison, secondary extinctions (ex mark) occur instantaneously. For a chain of three species, a background extinction of species B results in a secondary extinction of species A. Species B can recolonize and persists until species C suffers a background extinction. The network then collapses until species C colonizes. With C present, B can recolonize, which makes it possible for A to colonize. However, if A then drives B extinct, only C remains. The time-weighted occupancy of each species divided by the total time of the simulation gives the probability of observing a species in a sample (listed to the right of the diagram)
There are many ways to assess the stability or persistence of ecological networks. An early approach, still in wide use today, is the analysis of various properties of a community matrix (Levins 1968). For instance, loop analysis takes estimates of direct interactions among species and uses matrix algebra to estimate how a community will respond to perturbations. A second approach, as implemented more recently for many-species systems, is simulation of food-web dynamics (Yodzis and Innes 1992). These models, which allow for effects of consumers on resources and vice versa, yield detailed information on biomass or population density over time for each species in the network (Chen and Cohen 2001; Brose et al. 2006; Berlow et al. 2009). Such models are high dimensional as they require a variety of parameters for vital rates, density dependence, functional responses, and contact rates. A third promising approach uses metacommunity analysis (Leibold et al. 2004, Holyoak et al. 2005, Amarasekare 2008). Instead of abundances, metacommunity models track either the proportion of patches occupied by each species in a network (Melian and Bascompte 2002), each of the 2n possible sub-networks that make up a network (Holt 1997), or each link in a network (Pillai et al. 2010), considering, for instance, whether these proportions are stable. So far, most metacommunity models have focused on simple networks within a patch (but see Fortuna and Bascompte 2006). A fourth approach, robustness analysis, also considers species presence-absence, but focuses on changes in network structure due to loss of species and the secondary extinctions that result from resource loss (Solé and Montoya 2001; Dunne et al. 2002). This topological approach to simulating disassembly of ecological communities requires few assumptions about how species interact, and thus allows analyses of complex species dependencies not amenable to dynamical modeling (Dunne and Williams 2009). SENO models borrow elements from all of these approaches to provide an additional, complementary tool for ecologists studying ecological networks.
SENO models: overview
Patch occupancy and species probabilities
The probability of a species being present in a sample (or occupying a patch) equals its prevalence among similar samples. For instance, niche distribution models such as GARP and MAXENT use associations between species records and habitat variables to generate maps that predict the chance that a species will occur at a particular location (Phillips et al. 2006). Layering niche maps of multiple species estimates the expected species richness at a particular location (Young et al. 2009).
Species probabilities are a key output of patch occupancy models (Hanski and Gilpin 1991). Patch occupancy models assume that characteristics of the species, the landscape, and the spatial scale affect colonization rates. Each species also suffers a background extinction rate (e.g., due to the environment, demography, or anthropogenic effects), which, like colonization, could depend on species traits and spatial scale. The distinguishing feature of a SENO model from a metapopulation model is that extinction rates also depend on a complex network of interactions with other species.
Resource requirements
Consumers need resources to persist. In a patch occupancy model, a consumer requires concurrent occupancy of at least one resource. Generalist consumers are less likely to suffer from the loss of a resource if they are able to switch to, or increase reliance on, other resources (Solé and Montoya 2001; Dunne et al. 2002). Thus, highly connected networks filled with generalists are more robust to secondary extinction (Solé and Montoya 2001; Dunne et al. 2002). For the same reason, generalists should be able to colonize a wider range of patches.
Box 1. The challenge of modeling resource dependence
Mean-field approaches exist for handling resource dependencies in an occupancy model. The equilibrium model of island species richness (MacArthur and Wilson 1967) provides an example for occupancy, p i, of species i in a patch with a constant rate of colonization, c, from an outside region, and a probability of extinction within the patch, e, as δp i/δt = c(h − p i) − p i e, and p i* = h c / (c + e). Here, h might represent the probability that resources necessary for persistence are present. If resources are substitutable and independently distributed, h = 1 − Π(1 − p j), where p j is one of potentially several substitutable resources. For example, imagine a carnivore that feeds on two herbivores, each of which, at equilibrium, has a 0.25 probability of occurring in a patch. Assuming the occupancies of the herbivores are independent of each other, the probability, h, of at least one herbivore being present in a patch = 0.44.
However, in ecological networks, species occupancies are often not independent. In the above example, imagine that both herbivores specialize on a plant that has a 0.75 probability of occurring in a patch (i.e., for the herbivores, h = 0.75). Given p i* = 0.25 for each herbivore, and 0 possibility of occupying the patches without their host plant, each herbivore must have a 0.33 occupancy in patches with the plant. Their co-dependency on a shared resource leads to a positive correlation between the herbivores among patches. Due to this correlation, the probability of at least one herbivore occupying a patch is 0.5(1 − [1 − 0.33]2) = 0.275, less than the 0.44 expected by independent assortment. This is a simple example of using Bayesian network analysis (Jensen 1996) to solve conditional probabilities in a food web. Though Bayesian networks are ideal for modeling resource dependencies in a food web, they can only solve directed acyclic graphs, a property violated by consumer effects on resources (see below). Stochastic occupancy models can simulate how resource dependencies influence species occupancy. With enough replication, the average of a stochastic model converges on the expected probabilities of a Bayesian network, but does not require the constraining assumption that consumers cannot affect resources. Stochastic occupancy models are relatively easy to program and have a history in the ecological literature (Lande et al. 2003). END OF BOX
Although they have not been described as such, most robustness analyses are specialized stochastic occupancy models (specialized in that they lack colonization events and they remove a single species at each time step). A SENO model extends robustness analysis so species can colonize patches and extirpate other species. In addition, SENO models track transitions in continuous time instead of removing species as a series of inevitable events. One measure of network robustness to secondary extinction is the proportion of species that need to be removed to reduce species richness by some percentage, such as 50% (Dunne et al. 2002; Srinivasan et al. 2007). To measure robustness in a SENO model, one could first estimate the conditions (e.g., spatial scale, see below) at which expected species richness in a network of S species falls to 50% and then determine the expected species richness at this spatial scale in the absence of species interactions (see the “Methods” section), perhaps decomposing bottom-up and top-down effects.
Consumer pressure
A consumer can drive a preferred resource species to extinction if the resource lacks refuge from the consumer (Gonzalez-Olivares and Ramos-Jiliberto 2003). Such “top-down” extinctions have been demonstrated experimentally (Schmitz 2003), and can occur on islands after invasion (Hadfield et al. 1993). For this reason, enemy-free space is an important element of the realized niche (Hopkins and Dixon 1997). Furthermore, impacts on shared resources are the basis for interspecific competition, which also constrains the realized niche (Guisan and Thuiller 2005). Strong effects of consumers are ubiquitous in ecological systems: crows can eliminate wood pigeons (Tomialojc 1978); nest predation by skunks strongly depresses waterfowl abundance (Greenwood 1986); and sea lamprey nearly extirpated lake trout from the Great Lakes (Mills et al. 2003). These consumer-mediated direct effects can indirectly alter ecological network structure and dynamics: dingoes can greatly reduce red kangaroo populations, altering plant communities (Caughley et al. 1980); sea stars exclude mussels from the lower intertidal, permitting colonization of fewer dominant space holders (Paine 1966); and myxoma virus reduces rabbit abundance and changes plant communities (Fenner and Ratcliffe 1965). To incorporate such consumer-resource dynamics, ecologists have often used two-species consumer-resource models (Lotka 1925), or three- to four-species community modules (Holt 1997; McCann and Hastings 1997). However, these same species, when analyzed in a complex network (Chen and Cohen 2001; Brose et al. 2006; Berlow et al. 2009) might have opposite associations due to competition and other indirect effects (Yodzis 1998). Adding consumer effects allows SENO models to address trophic cascades, apparent competition, and competition for shared resources.
How might consumer effects vary? Predators have higher impacts per prey when they feed on fewer prey species (Edwards et al. 2010). This could occur if specialist consumers are more efficient at tracking and consuming their resources (e.g., Yamada and Boulding 1998), leading to a tradeoff between generality and impacts on resources that is akin to the “jack of all trades, master of none” concept (MacArthur 1972). Some parasitic organisms (particularly parasitoids and parasitic castrators) can strongly affect their hosts (Lafferty and Kuris 2009), and there are conditions under which parasites can cause host extinction (de Castro and Bolker 2005). However, most parasites are self-limiting within a consumer, making it less likely that they would cause extinction. For instance, crowding within a host can strongly limit intestinal worms (Read 1951), and endothermic hosts usually clear pathogens and gain permanent immunity, limiting the effect of pathogens at the population level (Norman et al. 1994).
Box 2. Modeling consumer effects
A food web can be represented by a matrix, L, where the column and row headings are the species list and an entry of 1 in cell L ij indicates that the species in column j feeds on the species in row i, whereas a 0 in cell L ij indicates no interaction. In a SENO model, we construct an additional matrix, K, that considers the effect of feeding links as a rate such that K ij represents the rate that a consumer, j, will extirpate a resource.
Incorporating the extent of competitive exclusion among basal species requires a pair of matrices. M ij is an estimate of resource overlap (from 0 to 1) between species i and species j (MacArthur and Levins 1967). Because competitors can differ in their resource use, M ij is not necessarily the same as M ji. For instance, a species with broad resource requirements can entirely overlap with a specialist but not vice versa. In addition, D ij is the proportion of the overlap in resources between species j and species i that is obtained by species j (and D ji + D ij = 1). A measure of the rate that species j will competitively exclude species i is, therefore, D ij M ij. For D ij = 0.5 (our assumption later), species are equal in their competitive abilities. Although we focus on basal taxa, competition could occur among consumers of resources that are not explicitly identified in a network (e.g., detritus, space on a substrate, nesting sites).
Scale, parameters, and expected species richness in a sub-network
All ecological networks have an implicit spatial scale within which the member species interact (McCann et al. 2005). For instance, empirical trophic networks have been constructed for large areas such as the North East Atlantic Shelf (Link 2002) or small habitats such as bromeliads (Starzomski et al. 2010). Not surprisingly, aspects of network structure such as species and link richness and other raw property values can change when viewed at different spatial and temporal scales. For instance, Warren (1989) found differing sub-networks as a result of sampling effects, habitat heterogeneity, and seasonality. He constructed a 36-species food web for Skipwith pond, but, by sampling at finer spatial scales (five sweep nets, monthly on both sides of the pond) found sub-networks of 12–32 species that varied over time. These sub-networks could be further divided into sub-networks associated with a depauperate open water habitat and a rich pond margin habitat. Species interactions like those in samples from Skipwith pond can scale up from simple community modules (Holt 1997) to complete food webs that persist over larger scales (Pillai et al. 2010; Stouffer and Bascompte 2010). For this reason, all else being equal, larger systems generate richer ecological networks. Larger lakes (Post et al. 2000) and islands (Takimoto et al. 2008) can also have longer food chains.
Rosenzweig (1995) gives two reasons why species richness increases with spatial scale. First, a sampling effect occurs within a patch because the higher a species’ density, and the greater the area under investigation, the higher the probability the species will occur in the sample. Second, the larger the sample, the greater the opportunity for habitat heterogeneity or limited dispersal within the sample (Shen et al. 2009). The increase in complexity in networks at larger spatial scales is not simply the result of combining species–area curves among taxa. In particular, upper trophic levels rely on the presence of their resources and this dependency increases the slope of the species–area relationship for species at higher trophic levels (Holt et al. 1999). Large predators are doubly sensitive to spatial scale due to their body size and dependency on resources that are more likely present in larger patches (Srivastava et al. 2008).
Species transition rates should be a function of spatial scale, and this may interact with body size. Extinction rate might decrease monotonically with patch area, whereas colonization rate might increase linearly (Hanski 2008). Empirical observations of metapopulations indicate that the probability of extirpation is higher in smaller patches (e.g., Lafferty et al. 1999). The way species vary with spatial scale likely changes with body size because larger species are less densely aggregated and require greater minimum areas to persist (Damuth 1981). The extent that competitive exclusion occurs could also change with scale. Resource overlap should decrease with spatial scale if larger areas have more habitat types, increasing the partitioning of otherwise limiting resources (Rosenzweig 1995). Furthermore, smaller organisms interact with a finer-grained environment and should find a particular area more heterogeneous than would large organisms (Kotliar and Wiens 1990). Direct consumer effects could also change with spatial scale, body size, and aspects of the consumer-resource interaction. Enemy-free space includes portions of the habitat that are not available to consumers, and increasing spatial scale elevates the chance that a sample will include some enemy-free space (Comins et al. 1992). For these reasons, if a consumer and resource co-occur, the probability of the consumer driving the resource extinct should decrease at larger spatial scales (Holyoak and Lawler 1996; Warren 1996). Larger consumers have greater energetic demands and, all things being equal, have greater per-capita capacity for consumption (Brown 1995). Therefore, the effect of a consumer might increase with its relative body size (Reuman and Cohen 2005). Finally, larger resources might require larger and thus less-common refuges, increasing their susceptibility to consumers at smaller spatial scales. Appendix 1 provides an example of how the probability of finding a particular species in a network could increase with spatial scale and how this scaling might differ with body size.
In short, because spatial scale matters for ecological networks, transition rates in SENO models should be scalable. Appendix 1 indicates how we derived scaling functions using metapopulation theory and other logical relationships. In the results, we explore in detail how making rates a function of spatial scale greatly affects model outputs.
SENO models: example
Model structure
In technical jargon, a SENO model is a depth-first search embedded in a first-order continuous-time Markov process applied to species occupancy in an ecological network. A species vector tracks occupancy with a 1 indicating a species is present or a 0 indicating a species is absent. Over time, species can transition between being present or absent. Background extinction rates represent species-specific environmental, demographic, or anthropogenic effects that lead to species loss. Colonization is the rate of entry into the patch or sample area. For simplicity, we assumed colonists arrive at a constant, species-specific rate from a larger, unspecified location. Up to this point, we have a multi-species stochastic patch occupancy model for a single island near a mainland (Appendix 1).
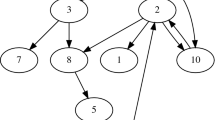
To accommodate species interactions requires an empirical or theoretical food web. For illustration purposes, we used the niche model (Williams and Martinez 2000) to generate a simple ecological network with three basal species and seven consumer species, coded in order of their position along the niche axis. This network had 17 directed trophic links (Fig. 2), represented as an adjacency matrix with a row and column for each species, 1 representing the column species feeding on the row species and 0 representing the absence of a link. For every link between a consumer and a resource in the adjacency matrix, there was rate of consumer-driven extinction for the resource (Box 2). Because basal nodes do not have explicit resources (e.g., no link to water or nutrients) in most topological networks, there is no way for them to deplete these resources and compete. Our indirect solution was to allow interaction among basal species (Box 2). Rates of competitive exclusion among basal species were as in Appendix 1.
Graphical representation of an example ecological network used in the analyses. Image produced with Network3D software written by R.J. Williams and available from the Pacific Ecoinformatics and Computational Ecology Lab, www.foodwebs.org. The network was randomly created by the niche model (Williams and Martinez 2000) and has three basal, seven consumer species, and 17 links. Species are spheres whose volume represents probability of occurrence from the intermediate spatial scale (A = 25). Links are tapering lines with the wide end coming from the consumer and tapering toward the resource. The list of links (consumer, resources) is: [(3, 1), (6, 1), (9, 1), (10, 2), (10, 3), (10, 6), (10, 4), (10, 8), (10, 5), (10, 7), (4, 1), (8, 2), (8, 3), (8, 1), (7, 2), (7, 3), (7, 4)], vertical height and shading represent weighted trophic level. Number codes are in order of body size, 1 being smallest
Species transition rates (colonization, background extinction, consumer exclusion, competitive exclusion of basal species) can be functions of spatial scale, the body size of the focal species, and characteristics of other species directly linked to the focal species, such as their body size, relationship with the focal species (consumer, resource or, for basal species, competitor), and strength of interaction with the focal species (weak or strong consumer pressure and basal competition). Appendix 1 describes the rate functions we used. Rates, transitions, and species comprise a rate table that sets the rules for the simulation. During a simulation, many potential transitions are irrelevant and it increases efficiency to ignore them. Specifically, we updated the table between transitions to exclude colonization for species currently present and exclude extinction related to any species currently absent. One could also update the table between transitions to simulate disturbances, climate change, restoration, controlled extirpations, etc. For instance, if climate affected the probability of extinction and climate changed over time, background rates of extinction could then change with time and be matched to the time of each transition (so long as transition rates were more frequent than changes in climate).
An iteration of a SENO model comprises a transition followed by secondary extinctions. Randomly sampling from the exponential distribution of the transition rates generates expected waiting times for the possible transitions from which the transition with the shortest wait time is selected. After implementing the selected transition, we updated the network and then retained only those consumers with resources.
There are several ways stochastic occupancy models can incorporate secondary extinctions. Computer programs can iteratively solve secondary extinctions, but this can be inefficient for large networks. A more sophisticated approach from graph theory is to use a depth-first search of the rooted network (Allesina et al. 2005). Rooting a network involves assigning a basal species as a root node as a resource (Allesina et al. 2009). For instance, sunshine is a reasonable root of many ecological networks. A depth-first search starts at the root and continues up each chain until it dead-ends at a node (Tarjan 1974). Breaks in a chain due to an extinction can leave higher trophic levels without a chain to the root. Likewise, colonists that cannot link to a chain connected to the root will not be able to persist. Therefore, only species accessed by a depth-first search will persist after a transition. This process must be recursive if species have multiple life history stages because the loss of one stage necessarily leads to the loss of all other stages.
If the transition was a colonization, the depth-first search prevented colonists from establishing if they lacked resources. If the transition was an extinction, the depth-first search eliminated species in chains wholly dependent on the lost species. We saved the resulting species vector and point in time as a record of the iteration before calculating a fresh set of waiting times to repeat the iteration.
We tracked the presence and absence of each species for 10,050 transitions. We chose to delete the first 50 transitions from the analyses to reduce the influence of starting the simulation with a complete network. Species probabilities were the proportion of time a species was present in a simulation (relative frequency weighted by time interval present). The supplementary material provides an annotated Mathematica™ demonstration that can be run without a Mathematica™ license.
Questions investigated
What questions can SENO models address? Comparisons could be made among networks, such as investigating the association between network complexity (richness and connectance) and measures of stability (such as robustness), but that is outside the scope of this paper. To illustrate how a SENO model can investigate species occupancies within a network, we considered: (1) the distribution of sub-network types, (2) the influence of spatial scale and body size, (3) robustness to species interactions, and (4) species interactions across spatial scales (either as input, as revealed by species removals, or as would be observed from correlations). We note that this effort is intended to show the utility of SENO models more than to test novel hypotheses.
To graphically illustrate the sub-networks that result from a SENO model, we saved an example of the species vector for 200 transitions at the intermediate spatial scale (A = 25). For the small, intermediate and large spatial scales, we also counted the number of unique sub-network types that occurred in 10,000 transitions, noting which types were disproportionately common in a simulation.
We then investigated two basic patterns from ecology that we assumed would arise from the models: a body size–abundance curve and a species–area curve. We plotted the probability of each species against its body size (100n, where n was the niche value of the species in the niche model) at three spatial scales (area = 2.5, 25, 250) to test the expectation that larger species would be less frequently observed, particularly at small spatial scales. To evaluate the assumption that richness would increase with spatial scale, we conducted simulations across a range of scales, and plotted the probability of each species as a function of spatial scale (logged over four orders of magnitude, A = 1 to 1,000), generating a stacked species–area curve.
To estimate the robustness of the ten species network to species interactions, we determined the spatial scale at which species richness was 50% of the original network (A 50) so we could estimate network robustness. For a comparative estimate of the background expected species richness in the absence of species interactions at A 50, we used \( {S_b} = \Sigma \left[ {1 - {E_b}/\left( {{E_b} + C} \right)} \right] \), where C and E b are rates of colonization and background extinction at A 50. Robustness, R, to species interactions in general (not just secondary extinction due to resource loss) was, therefore, R = (S – S b) / S, and a network is completely robust to species interactions if R = 0.5.
We investigated interactions and associations among species in the network at different spatial scales (Area = 2.5, 25, 250). This was done in three ways. First, the direct effect of one species on the occupancy of another was predicted from interactions between species as specified from the network structure and model inputs. Here, the effect of a resource on a consumer was 1 / g where g was the generality of the consumer. The rate a consumer extirpated a resource or one basal species extirpated another was standardized to a value between 0 and 1, using the cumulative distribution function of the exponential distribution of the corresponding rate (for T = 1) in the rate table. Second, to determine the effect of one species on another in the context of the network (e.g., the combined action of direct and indirect effects), we enacted individual species removal experiments. Each trial excluded a single species after which a simulation was run and expected species richness of a sub-network calculated. Here, the value of a cell also corresponded to the standardized effect of the species in the column on the species in the row. Specifically, the probability of occurrence of each species in a row, P r, was calculated for the complete network. That species’ probability was then recalculated after removing the species in the column from the network, (P r-c). The effect of the column species on the row species was standardized between 1 and −1, as (P r − P r-c) / P r, for P r > P r-c, otherwise (P r − P r-c) / P r-c, providing a measure, for each cell, similar in principle and scale to a correlation coefficient. Third, to determine associations among species as might be observed in samples from nature, we generated a phi correlation matrix among species across the sub-networks resulting from the simulations.
We compared the direct and indirect effects of a species on the expected species richness of a sub-network (for A = 25). We estimated expected species richness from simulations where we removed a target species, c, (∑P –c). The direct contribution of species, c, to the expected species richness of a sub-network was simply its probability of occurrence, P c. The effect of species c on the other species in a sub-network was ∑P − ∑P –c. We then plotted each species’ direct vs. indirect contribution to expected species richness of a sub-network, noting which species facilitated expected species richness of other species, which species impacted less than one other species when present (therefore contributing to net species richness), and which species led to a net loss of expected species richness because they impacted more than one other species when present.
Results
Although we used a single network to illustrate SENO models, the general conclusions are robust to a variety of networks. Increasing background extinction, basal competition, and consumer effects decreased expected species richness in a sub-network. At small spatial scales, basal competition and the background rate of extinction were high (particularly for large species), and consumer impacts on resources were high, (particularly for large consumers).
Simulations produced many sub-network types. As an example of the non-equilibrial dynamics produced by a simulation, we present a segment of the time series for spatial scale A = 25. Even for this small window of transitions, richness in a patch ranged from zero to nine species (Fig. 3). For the ten-species network, there are up to 210 possible topologies (or sub-networks). The distribution of sub-network types varied with spatial scale, with the intermediate scale having the greatest diversity of sub-network types (Table 1).
The sequence of state transitions numbered 1,001 to 1,200 out of a 10,000-transition simulation of the ten-species network in Fig. 2. The area between horizontal lines represents a species (i.e., species are stacked with species 1 at the bottom and species 10 on the top). Dark fill indicates species occupancy in the food web. Transition sequence (not time) is along the horizontal axis. The right vertical axis label is the prevalence of each species in the sample (probability of occurrence), which is the relative frequency of each species among the transitions weighted by the duration of the interval between transitions (as in Fig. 1)
Spatial scale and body size affected the results. A strong negative association between body size and species prevalence occurred, and the slope of this relationship was steepest at the intermediate scale (Fig. 4). The expected species richness in a sub-network increased with spatial scale as one would expect from a species–area curve with larger species being disproportionately missing at smaller spatial scales (Fig. 5). Expected species richness was 5 (50% of total) at a spatial scale of A = 14.1. For A = 14.1, expected species richness after ignoring species interactions (\( \Sigma \left[ {1 - {E_b}/\left( {{E_b} + C} \right)} \right] \) was 6.5. Therefore, in our example network, robustness to species interactions was 0.45 (out of 0.5).
Decrease in species prevalence (probability of occurrence) with body size for the ten species network at three spatial scales. Rates of background extinction, basal competition, and consumer effects were all scaled with body size according to Appendix 1
Increase in expected species richness in a sub-network with spatial scale for the ten-species network in Fig. 2. Species are stacked with species 1 at the bottom and species 10 on the top. Rates of background extinction, basal competition, and consumer effects were all scaled with body size according to Appendix 1
Inputs to the model suggested certain types of associations, but indirect effects altered how species affected each other. Row 1 of Fig. 6 illustrates how the inputs to the model would have predicted species interactions. There were expected positive effects of basal species on consumers, competition among basal species, negative effects of consumers on resources (which decreased with spatial scale) and positive effects of resources on consumers. The species removal experiments indicated that indirect effects led to different patterns from the inputs (Fig. 6 row 2). For instance, at the smallest spatial scale, some expected strong consumer effects were weak because the consumers infrequently occurred in a sub-network. In addition, expected neutral interactions among species were sometimes negative due to indirect effects such as competition among consumers. At larger spatial scales, consumer effects on resources were less important, and resource control dominated interactions. Correlations among species in the outputs of the simulations did not match the inputs or the results of the species removals. Most correlations among species were relatively weak (Fig. 6 row 3). Only a few species pairs were negatively correlated and the few strong correlations were positive, suggesting resource control was greater than consumer control. Correlations increased in strength at larger spatial scales because a reduction in consumer impacts left only positive associations between resources and consumers.
Interaction maps of species associations and effects for the ten-species network in Fig. 2. Species codes (1–10) represented along the top and right sides. Columns represent spatial scale from small to large. Shading represents the strength of the association between two species, with gray being neutral, black positive and white negative: first row interactions between species as input into the model. Second row standardized effect calculated by removing the column species and noting the standardized change in the row species (i.e., white indicates that the column species impacted the row species), including the sum of direct and indirect effects. Third row map of phi correlations among species as determined from observations of the network over time
To investigate how each species affected the network as a whole, we plotted direct versus indirect contributions to expected species richness for the ten species (A = 25). This approach indicated how species could facilitate, contribute to, or reduce expected species richness (Fig. 7). The two widely consumed basal species (1 and 2) had an indirect positive contribution to the probability of other species occurring in a sub-network. We refer to such species as “biodiversity facilitators”. Species 1 might even qualify as a foundational species due to its strong positive effect on expected species richness in a sub-network. Biodiversity facilitators tended to be small-bodied species fed on by many consumers. Most species had slightly negative effects on the other species in a sub-network, but, because the expected richness of the sub-network was still greater in their presence, we refer to them as “biodiversity contributors.” Species 10 reduced biodiversity. We therefore considered this large top predator a “biodiversity inhibitor”.
Individual species effects on expected species richness in a sub-network. The effect of each species on expected species richness is parsed into the probability that a species occurs (horizontal axis) and the indirect effect of the species on the other members of the sub-network (vertical axis). Numbers indicate species codes from Fig. 2. Two lines divide the space into three qualitative outcomes. Biodiversity facilitators have a net positive indirect effect on other species. Biodiversity contributors have a net negative indirect effect on other species, but their own presence (their direct effect) outweighs this negative effect. Biodiversity inhibitors are species that have stronger net indirect effects than direct effects. Lower expected species richness occurs with the presence of biodiversity inhibitors and greater expected species richness occurs with the presence of biodiversity facilitators and contributors
Discussion
Stochastic ecological network occupancy models share elements of several past approaches. Like loop analysis, SENO models require an interaction matrix. They are similar to population dynamics models in their ability to account for consumer effects and basal competition. However, like patch models and robustness analyses, SENO models track occupancy, not density. A SENO model is also like a robustness analysis in its ability to handle complex networks and its reliance on stochastic modeling. Like a Bayesian network analysis, a SENO model solves the challenge of conditional probabilities, but through replication and averaging instead of algorithms. Like metapopulation models, SENO models allow transition rates to vary with spatial scale.
SENO models output a series of sub-networks and their durations, helping to visualize temporal dynamics and identify persistent sub-networks. Averaging sub-networks (weighted by their durations) estimates the probability of species occurrences. Manipulating inputs helps to estimate robustness of the network, describe species–area relationships, and identify the roles of individual species in a network. However, SENO models do not include other information of interest to ecologists such as system-wide stability criteria, densities, biomass, and consumption rates. Thus, SENO models are on a continuum between the high-input requirements and rich yield of dynamic models and lower input requirements and limited yield of traditional robustness analysis. We see SENO models as complementing, not supplanting, existing approaches for analyzing ecological networks.
We used a simple food web to illustrate SENO models, but additional biological detail is easy to add. A SENO model on a personal computer can analyze relatively complex networks with hundreds of species (e.g., 10,000 iterations of a 100-species network presently take 1–15 min.). Minor modifications to our example could allow several additional types of complexity. Non-substitutable resources (i.e., to persist, a species must have resources A and B, not A or B) could represent life stages with different resource requirements. Dependency loops between species that do not deplete each other could indicate mutualisms (e.g., pollination). Resources that consumers cannot extirpate could represent subsidies that lead to donor control of consumers (Polis et al. 1997). Non-substitutable resources that are not subject to extirpation by consumers could specify non-trophic dependencies, like species that provide habitat (e.g., hosts to commensals, trees that provide nest sites for birds). Also, specifying habitat types as nodes could allow more explicit consideration of habitat dependencies. Finally, instead of colonists coming from a regional pool (our island-mainland model), patches could be linked to allow for metacommunity dynamics. SENO models could occur within a network of patches, for instance, by embedding them within a stochastic patch occupancy model. Several other system or question-specific alterations are possible.
Although many studies have considered how food-web structure changes (or does not change) with increasing species richness (Dunne 2006), few have explicitly explored if and how the structure of ecological networks changes with spatial scale (but see Martinez and Lawton 1995; McCann et al. 2005; Pillai et al. 2010). Given the assumption that background, resource, and consumer effects decrease with scale, the direct effect of a species on the expected species richness in a sub-network approaches unity with increasing spatial scale, whereas the indirect effect declines to zero for most species. This suggests that SENO models can indicate how predictions for some ecological hypotheses will vary with the scale of observation. For instance, this spatial scaling phenomenon could help resolve a conflict in the literature surrounding the effects of invasive species. Although most conservation biologists argue that control of invasive species protects biodiversity, invasive species can contribute to species richness because their addition to regional faunas offsets their indirect impacts on native species (Sax et al. 2002). SENO models might indicate that impacts of invasive species on expected species richness are likely to be scale-dependent, with strong negative effects (biodiversity inhibition) possible on local scales but positive effects (biodiversity contribution), such as measured by Sax et al. (2002), at regional scales (as happens for species 10 in our example).
SENO models might help generate spatial maps of networks based on species distributions. For instance, applying habitat suitability maps for all species in a region along with their potential links creates the possibility of generating spatially explicit SENO models where the background probability of extinction for each species corresponds to the suitability of the habitat at that point. SENO models could also be used to explore temporal patterns, which could be useful for studying restoration and resilience.
The sub-network types in our simulations suggested the network was a mosaic comprised mainly of a few common sub-networks (Kondoh 2008) and several rare sub-networks. For instance, a food web of Costa Rican bromeliad habitats has 70 invertebrate species but sub-networks in individual bromeliad plants typically have five to 20 species (Starzomski et al. 2010). Much work has been done on the dynamics of small networks such as tri-trophic food chains and slightly more complex modules that incorporate omnivory, exploitative competition, and apparent competition, but less is known about how these modules combine to form larger networks (Stouffer and Bascompte 2010). One idea is that food webs might be formed primarily from inherently stable modules, but less stable modules can also persist when they are integrated into webs in ways that are stabilizing (Kondoh 2008). Another view is that mobile predators couple sub-networks as they move across a landscape (McCann et al. 2005). SENO models can help address these hypotheses by indicating which community modules represented in a network are most likely to persist as sub-networks. In our example, sub-networks were not random subsets of the parent network: they clustered, and a few topologies were both frequent and durable (see Allesina and Pascual 2008). Clusters of similar sub-networks might represent alternate states, and it seems possible that the colonization or extinction of key species might result in rapid wholesale shifts in the community toward a different cluster of sub-network types.
SENO models are a convenient way to make effect maps, like Fig. 6, for illustrating the roles of species in ecological networks. Inputs to the model did not directly predict how species interacted, suggesting that complex indirect effects can overwhelm direct effects in food webs. This, of course, is a main argument for studying species interactions in the context of ecological networks. Correlations among species (such as the observations ecologists commonly make) did not match interactions among species for two reasons. First, some associations might be non-causal. For instance, variation in a predator could lead to positive correlations among prey. Second, opposing effects cancel each other, obscuring how a resource can benefit a consumer but also be depressed by that consumer.
SENO models allow the categorization of species by their combined effects on the network. The classification of biodiversity facilitators, contributors, and inhibitors might be useful for determining which native species should be targeted for preservation (e.g., facilitators), and which introduced species should be targeted for control (e.g., inhibitors). We suspect that this might lead in some cases to counterintuitive results. For instance, do large predators tend to be facilitators, contributors, or inhibitors of biodiversity? How about parasites? Does this change with spatial scale?
SENO models can estimate network metrics such as trophic level and the related concept of food-chain length, a topic of abiding interest for ecologists (Post 2002a, b; Williams and Martinez 2004) and with important implications for bioaccumulation (Kelly et al. 2007). The trophic level of a species is a measure of how many times chemical energy is converted into biomass via trophic interactions. By convention, primary producers are assigned a trophic level of 1. However, the trophic levels of other taxa are often less clear, as there can be many chains of different lengths that connect consumers to primary producers (i.e., omnivory) with additional complications arising from loops such as cannibalism, mutual predation, and longer cycles. Also, parasitic or mutualistic carbon transfers between plants, for example via mycorrhizae (Simard et al. 1997), can obscure the trophic level of primary producers, and the trophic level of individuals within a population or species can vary greatly. Species’ trophic levels have been estimated in a variety of ways, primarily using network structure with or without energy flow information (Post 2002a; Williams and Martinez 2004), or using stable isotopes of carbon and nitrogen (Post 2002b). Each of the approaches requires a different type of data and assumptions, and thus they result in various estimates of trophic level. SENO models can provide an alternative method for estimating trophic levels by weighting the contribution of each resource in the diet of a consumer by the probability that the resource occurs in a sub-network. Variation in trophic level among sub-networks could also indicate the extent to which the trophic level of a species might vary across a landscape or over time. It would be informative to cross-validate stable isotope, network structure, and energy flow information with SENO model estimates of trophic level.
SENO models could easily explore general questions using theoretical networks but can they be applied to empirical networks? SENO models, like most network models, require a large number of biological assumptions that will be difficult to estimate from field data without resulting to short cuts such as allometric scaling as we have in this paper. That said, general patterns available from field data, like species–area curves or changes in occupancy with spatial scale and body size, could be compared to outputs of SENO models. SENO models could also help make theoretical predictions about removal and exclusion experiments. Their outputs could be compared with real data from microcosm experiments and networks with known species occupancies. However, evaluation of SENO models with empirical data will be challenging.
We have illustrated how to construct SENO models and suggested several analyses at the level of the network and of the individual species. Our simple examples suggest that SENO models can produce results consistent with basic ecological concepts such as species–area curves and body size–abundance associations. Future work could consider the relationship between robustness and complexity in SENO models or could examine the contribution of species interactions to species–area curves. From an applied perspective, SENO models could help identify characteristics of invasive species that reduce biodiversity or of foundational species that would foster the restoration of biodiversity. They could also identify key resource needs, competitors, and predators for endangered species or species that humans extract, and could estimate the community-wide effects of events such as over-fishing or species introductions. Our hope is that SENO models will provide a useful way to integrate ecological network research with spatial niche modeling for the support of ecosystem-based management. They potentially even have application in other disciplines that study networks such as economics, computer science, systems biology, and sociology.
References
Allesina S, Pascual M (2008) Network structure, predator-prey modules, and stability in large food webs. Theor Ecol 1:55–64
Allesina S, Bodini A, Bondavalli C (2005) Ecological subsystems via graph theory: the role of strongly connected components. Oikos 110:164–176
Allesina S, Bodini A, Pascual M (2009) Functional links and robustness in food webs. Phil Trans Roy Soc B 364(1524):1701-1709
Amarasekare P (2008) Spatial dynamics of keystone predation. J Anim Ecol 77:1306–1315
Bascompte J (2009) Disentangling the web of life. Science 325:416–419
Berlow EL, Dunne JA, Martinez ND, Stark PB, Williams RJ, Brose U (2009) Simple prediction of interaction strengths in complex food webs. Proc Natl Acad Sci 106:187–191
Brose U, Williams RJ, Martinez ND (2006) Allometric scaling enhances stability in complex food webs. Ecol Lett 9:1228–1236
Brown JH (1995) Macroecology. University of Chicago Press, Chicago
Caughley G, Grigg GC, Caughley J, Hill GJE (1980) Does dingo predation control densities of kangaroos and emus? Aust Wildl Res 7:1–12
Chen X, Cohen JE (2001) Transient dynamics and food web complexity in the Lotka–Volterra cascade model. Proc R Soc Lond B 268:869–877
Comins HN, Hassell MP, May RM (1992) The spatial dynamics of host parasitoid systems. J Anim Ecol 61:735–748
Damuth J (1981) Population-density and body size in mammals. Nature 290:699–700
de Castro F, Bolker B (2005) Mechanisms of disease-induced extinction. Ecol Lett 8:117–126
Dunne JA (2006) The network structure of food webs. In: Pascual M, Dunne JA (eds) Ecological networks: linking structure to dynamics in food webs. Oxford University Press, New York, pp 27–86
Dunne JA, Williams RJ (2009) Cascading extinctions and community collapse in model food webs. Philos Trans R Soc, Biol 364:1711–1723
Dunne JA, Williams RJ, Martinez ND (2002) Network structure and biodiversity loss in food webs: robustness increases with connectance. Ecology Letters 5(4):558
Edwards KF, Aquilino KM, Best RJ, Sellheim KL, Stachowicz JJ (2010) Prey diversity is associated with weaker consumer effects in a meta-analysis of benthic marine experiments. Ecol Lett 13:194–201
Fenner F, Ratcliffe FN (1965) Myxomatosis. Cambridge University Press, Cambridge
Fortuna MA, Bascompte J (2006) Habitat loss and the structure of plant-animal mutualistic networks. Ecol Lett 9:278–283
Gonzalez-Olivares E, Ramos-Jiliberto R (2003) Dynamic consequences of prey refuges in a simple model system: more prey, fewer predators and enhanced stability. Ecol Model 166:135–146
Greenwood RJ (1986) Influence of striped skunk removal on upland duck nest success in North Dakota. Wildl Soc Bull 14:6–11
Guisan A, Thuiller W (2005) Predicting species distribution: offering more than simple habitat models. Ecol Lett 8:993–1009
Hadfield MG, Miller SE, Carwile AH (1993) The decimation of endemic Hawaiian tree snails by alien predators. Am Zool 33:610–622
Hanski I (2008) Spatial patterns of coexistence of competing species in patchy habitat. Theor Ecol 1:29–43
Hanski I, Gilpin M (1991) Metapopulation dynamics—brief history and conceptual domain. Biol J Linn Soc 42:3–16
Holt RD (1997) Community modules. In: Gange A, Brown V (eds) Multitrophic interactions in terrestrial ecosystems, 36th symposium of the British ecological society. Blackwell, Oxford, pp 333–350
Holt RD, Lawton JH, Polis GA, Martinez ND (1999) Trophic rank and the species–area relationship. Ecology 80:1495–1504
Holyoak M, Lawler SP (1996) Persistence of an extinction-prone predator–prey interaction through metapopulation dynamics. Ecology 77:1867–1879
Holyoak M, Leibold MA, Holt RD (eds) (2005) Metacommunities: spatial dynamics and ecological communities. University of Chicago Press, Chicago
Hopkins GW, Dixon AFG (1997) Enemy-free space and the feeding niche of an aphid. Ecol Entomol 22:271–274
Jensen FV (1996) Introduction to Bayesian networks. Springer, Secaucus
Kelly BC, Ikonomou MG, Blair JD, Morin AE, Gobas FAPC (2007) Food web-specific biomagnification of persistent organic pollutants. Science 317:236–239
Kondoh M (2008) Building trophic modules into a persistent food web. Proc Natl Acad Sci USA 105:16631–16635
Kotliar NB, Wiens JA (1990) Multiple scales of patchiness and patch structure: a hierarchical framework for the study of heterogeneity. Oikos 59:253–260
Lafferty KD, Kuris AM (2009) Parasitic castration: the evolution and ecology of body snatchers. Trends Parasitol 25:564–572
Lafferty KD, Swift CC, Ambrose RF (1999) Extirpation and recolonization in a metapopulation of an endangered fish, the tidewater goby. Conserv Biol 13:1447–1453
Lande R, Engen S, Saether BE (2003) Stochastic population dynamics in ecology and conservation. Oxford University Press, Oxford
Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, Loreau M, Gonzalez A (2004) The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett 7:601–613
Levins R (1968) Evolution in changing environments: some theoretical explorations. Princeton University Press, Princeton
Link J (2002) Does food web theory work for marine ecosystems? Mar Ecol Prog Ser 230:1–9
Lotka AJ (1925) Elements of physical biology. Williams and Wilkins
MacArthur R, Levins R (1967) Limiting similarity convergence and divergence of coexisting species. Am Nat 101:377–385
MacArthur RH (1972) Geographical ecology. Patterns in the distribution of species. Harper and Row, New York
MacArthur RH, Wilson EO (1967) The theory of island biography. Princeton University Press, Princeton
Martinez ND, Lawton JH (1995) Scale and food-web structure—from local to global. Oikos 73:148–154
McCann K, Hastings A (1997) Re-evaluating the omnivory-stability relationship in food webs. Proc R Soc Lond B 264:1249–1254
McCann KS, Rasmussen JB, Umbanhower J (2005) The dynamics of spatially coupled food webs. Ecol Lett 8:513–523
Melian CJ, Bascompte J (2002) Food web structure and habitat loss. Ecol Lett 5:37–46
Mills EL, Casselman JM, Dermott R, Fitzsimons JD, Gal G, Holeck KT, Hoyle JA, Johannsson OE, Lantry BF, Makarewicz JC, Millard ES, Munawar IF, Munawar M, O'Gorman R, Owens RW, Rudstam LG, Schaner T, Stewart TJ (2003) Lake Ontario: food web dynamics in a changing ecosystem (1970–2000). Can J Fish Aquat Sci 60:471–490
Montoya JM, Pimm SL, Sole R (2006) Ecological networks and their fragility. Nature 442:259–264
Norman R, Begon M, Bowers RG (1994) The population dynamics of microparasites and vertebrate hosts: the importance of immunity and recovery. Theor Popul Biol 46:96–119
O'Gorman EJ, Emmerson M (2009) Perturbations to trophic interactions and the stability of complex food webs. Proc Natl Acad Sci 106:13393–13398
Paine RT (1966) Food web complexity and species diversity. Am Nat 100:65–75
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pillai P, Loreau M, Gonzales A (2010) A patch-dynamic framework for food web metacommunities. Theoretical Ecology. doi: 10.1007/s12080-009-0065-1
Polis GA, Anderson WB, Holt RD (1997) Toward an integration of landscape and food web ecology: the dynamics of spatially subsidized food webs. Annu Rev Ecol Syst 28:289–316
Post DM (2002a) The long and short of food-chain length. Trends Ecol Evol 17:269–277
Post DM (2002b) Using stable isotopes to estimate trophic position: models, methods and assumptions. Ecology 83:703–718
Post DM, Pace ML, Hairston NG (2000) Ecosystem size determines food-chain length in lakes. Nature 405:1047–1049
Read CP (1951) The crowding effect in tapeworm infections. J Parasitol 37:174–178
Reuman DC, Cohen JE (2005) Estimating relative energy fluxes using the food web, species abundance, and body size. Adv Ecol Res 36:137–182
Rosenzweig ML (1995) Species diversity in space and time. Cambridge Univ. Press, Cambridge
Sax DF, Gaines SD, Brown JH (2002) Species invasions exceed extinctions on islands worldwide: a comparative study of plants and birds. Am Nat 160:766–783
Schmitz OJ (2003) Top predator control of plant biodiversity and productivity in an old-field ecosystem. Ecol Lett 6:156–163
Shen G, Yu M, Hu X, Mi X, Ren H, Sun I, Ma K (2009) Species–area relationships explained by the joint effects of dispersal limitation and habitat heterogeneity. Ecology 90:3033–3041
Simard SW, Perry DA, Jones MD, Myrold DD, Durall DM, Molina R (1997) Net transfer of carbon between ectomycorrhizal tree species in the field. Nature 388:579–582
Solé RV, Montoya JM (2001) Complexity and fragility in ecological networks. Proc R Soc Lond B Biol Sci 268:2039–2045
Srinivasan UT, Dunne JA, Harte J, Martinez ND (2007) Response of complex food webs to realistic extinction sequences. Ecology 88:671–682
Srivastava DS, Trzcinski MK, Richardson BA, Gilbert B (2008) Why are predators more sensitive to habitat size than their prey? Insights from bromeliad insect food webs. Am Nat 172:761–771
Starzomski B, Suen D, Srivastava D (2010) Predation and facilitation determine chironomid emergence in a bromeliad-insect food web. Ecol Entomol 35:53–60
Stouffer DB, Bascompte J (2010) Understanding food-web persistence from local to global scales. Ecol Lett 13:154–161
Takimoto G, Spiller DA, Post DM (2008) Ecosystem size, but not disturbance, determines food-chain length on islands of the Bahamas. Ecology 89:3001–3007
Tarjan R (1974) Depth-first search and linear graph algorithms. SIAM J Comput 1:146–160
Tomialojc L (1978) Influence of predators on breeding woodpigeons in London parks. Bird Study 25:2–10
Warren PH (1989) Spatial and temporal variation in the structure of a freshwater food web. Oikos 55:299–311
Warren PH (1996) The effects of between-habitat dispersal rate on protist communities and metacommunities in microcosms at two spatial scales. Oecologia 105:132–140
Williams RJ, Martinez ND (2000) Simple rules yield complex food webs. Nature 404:180–183
Williams RJ, Martinez ND (2004) Limits to trophic levels and omnivory in complex food webs: theory and data. Am Nat 163:458–468
Yamada SB, Boulding EG (1998) Claw morphology, prey size selection and foraging efficiency in generalist and specialist shell-breaking crabs. J Exp Mar Biol Ecol 220:191–211
Yodzis P (1998) Local trophodynamics and the interaction of marine mammals and fisheries in the Benguela ecosystem. J Anim Ecol 67:635–658
Yodzis P, Innes S (1992) Body size and consumer-resource dynamics. Am Nat 139:1151–1175
Young BE, Franke I, Hernandez PA, Herzog SK, Paniagua L, Tovar C, Valqui T (2009) Using spatial models to predict areas of endemism and gaps in the protection of Andean slope birds. Auk 126:554–565
Acknowledgments
We appreciate input from S. Allesina, E. Baskerville, C. Briggs, A de Roos, G. DeLeo, A. Dobson, T. Gross, N. Martinez, J. McLaughlin, E. Mordecai, M. Pascual, S. Sokolo, D. Stouffer and T. Tinker. This work was conducted as a part of the Parasites and Food Webs Working Group supported by the National Center for Ecological Analysis and Synthesis, a Center funded by NSF (Grant #EF-0553768), the University of California, Santa Barbara, and the State of California. Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the US government.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Appendix 1
Scaling of ecological networks and adding biological complexity (DOC 30 kb)
This file is unfortunately not in the Publisher's archive anymore: NBP (DOC 61.9 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lafferty, K.D., Dunne, J.A. Stochastic ecological network occupancy (SENO) models: a new tool for modeling ecological networks across spatial scales. Theor Ecol 3, 123–135 (2010). https://doi.org/10.1007/s12080-010-0082-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12080-010-0082-0