Abstract
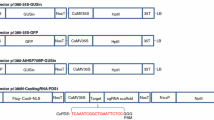
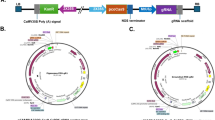
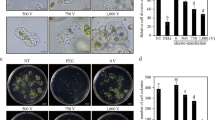
Citrus reticulata Blanco also known as kinnow mandarin is a widely grown horticultural crop in Punjab. CRISPR/Cas9 technology is being widely used for generation of varieties with increased resilience towards abiotic and biotic stresses as well as improved horticultural traits. Xanthomonas citri subsp. citri (Xcc)-mediated Agroinfiltration offers a fast and transgene-free method for the delivery of CRISPR/Cas9 constructs for systemic introduction into plants for functional genomics and expression studies. The technology is currently unexplored in kinnow mandarin. This study is aimed at establishing an efficient method of Cas9 delivery for transient knockout of PDS (phytoene desaturase) gene in kinnow mandarin. The construct pKO-119-PDS N-Cas9/sgRNA:PDS1 carrying sgRNA and Cas9 enzyme was delivered into the dorsal surface of young leaves of kinnow mandarin. The leaves showed albino patches at the point of injection within 60 h. Two surfactants (Triton-X and Silwet™) were used to ease the Agroinfiltration process which resulted in variation in the expression of vector. The Sanger’s analysis of the treated plants showed a substitution within the sgRNA region which resulted in change in amino acid from proline to serine. The protocol provides a feasible and an efficient method for genome editing in C. reticulata which could be helpful in future studies aimed at genome editing as well as genetic transformation.









Similar content being viewed by others
Data availability
Data available on request.
References
Ghosh, D., Venkataramani, P., Nandi, S., & Bhattacharjee, S. (2019). CRISPR-Cas9 a boon or bane: The bumpy road ahead to cancer therapeutics. Cancer Cell International, 19, 12. https://doi.org/10.1186/s12935-019-0726-0
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., & Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduction and Targeted Therapy, 5, 1. https://doi.org/10.1038/s41392-019-0089-y
Hoshijima, K., Jurynec, M. J., & Grunwald, D. J. (2016). Precise genome editing by homologous recombination. Methods in Cell Biology, 135, 121–147. https://doi.org/10.1016/bs.mcb.2016.04.008
Bernardi, B., & Wendland, J. (2020). Homologous recombination: A GRAS yeast genome editing tool. Fermentation, 6(2), 57. https://doi.org/10.3390/fermentation6020057
Chen, C., Fenk, L. A., & De Bono, M. (2013). Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acid Research, 41(20), e193. https://doi.org/10.1093/nar/gkt805
Carroll, D. (2011). Genome engineering with zinc-finger nucleases. Genetics, 188(4), 773–782. https://doi.org/10.1534/genetics.111.131433
Harmatz, P., Prada, C. E., Burton, B. K., Lau, H., Kessler, C. M., Cao, L., et al. (2022). First-in-human in vivo genome editing via AAV-zinc-finger nucleases for mucopolysachharides I/II and haemophilia B. Molecular Therapy, 30(12), 3587–3600. https://doi.org/10.1016/j.ymthe.2022.10.010
Wood, A. J., Lo, T. W., Zeitler, B., Pickle, C. S., Ralston, E. J., Lee, A. H., et al. (2011). Targeted genome editing across species using ZFNs and TALENs. Science, 333(6040), 307. https://doi.org/10.1126/science.1207773
Cerven, J., Vrbovsky, V., Horacek, J., Bartas, M., Endlova, L., Pecinka, P., et al. (2013). New low morphine opium poppy genotype obtained by TILLING approach. Plant, 12(5), 1077. https://doi.org/10.3390/plants12051077
Palan, B., Anjanabha, B., & Bharat, C. (2021). TILLING in the era of precise genome editing. Indian Journal of Biotechnology, 20(1), 9–16.
Acevedo-Garcia, J., Spencer, D., Thieron, H., Reinstader, A., Hammond-Kosack, K., Phillips, A. L., et al. (2016). mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnology Journal. https://doi.org/10.1111/pbi.12631
Luo, M., Li, H., Charkraborty, S., Morbitzer, R., Rinaldo, A., Upadhyaya, N., et al. (2019). Efficient TALEN-mediated gene editing in wheat. Plant Biotechnology Journal, 17(11), 2026–2028. https://doi.org/10.1111/pbi.13169
Kazama, T., Okuno, M., Watari, Y., Yanase, S., Koizuka, C., Tsuruta, Y., et al. (2019). Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nature Plants, 5, 722–730. https://doi.org/10.1038/s41477-019-0459-z
Boubakri, H. (2023). Recent progress in CRISPR/Cas9-based genome editing for enhancing plant disease resistance. Gene, 866, 147334. https://doi.org/10.1016/j.gene.2023.147334
Wada, N., Ueta, R., Osakabe, Y., & Osakabe, K. (2020). Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biology, 20, 234. https://doi.org/10.1186/s12870-020-02385-5
Samanta, M. K., Dey, A., & Gayen, S. (2016). CRISPR/Cas9: An advanced tool for editing plant genomes. Transgenic Research, 25, 561–573. https://doi.org/10.1007/s11248-016-9953-5
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., & Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Current Opinion in Biotechnology, 32, 76–84. https://doi.org/10.1016/j.copbio.2014.11.007
Danilo, B., Montes, E., Archambeau, H., Lode, M., Rousseau-Gueutin, M., Chevre, A. M., et al. (2021). I-SceI and customized meganucleases-mediated genome editing in tomato and oilseed rape. Transgenic Research, 31, 87–105. https://doi.org/10.1007/s11248-021-00287-2
Silva, G., Poirot, L., Galetto, R., Smith, J., Montoya, G., Duchateau, P., et al. (2011). Meganucleases and other tools for targeted genome engineering: Perspective and challenges for gene therapy. Current Gene Therapy, 11(1), 11–27. https://doi.org/10.2174/156652311794520111
Cox, D. B. T., Platt, R. J., & Zhang, F. (2015). Therapeutic genome editing: Prospects and challenges. Nature Medicine, 21(2), 121–131. https://doi.org/10.1038/nm.3793
Manchanda, P., & Suneja, Y. (2018). Genome editing for crop improvement: status and prospects. In S. S. Gosal & S. H. Wani (Eds.), Biotechnologies for crop improvement (Vol. 3, pp. 75–104). Berlin: Springer.
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Research, 23, 1229–1232. https://doi.org/10.1038/cr.2013.114
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nature Biotechnology, 31, 686–688. https://doi.org/10.1038/nbt.2650
Xie, K., & Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Molecular Plant, 6(6), 1975–1983. https://doi.org/10.1093/mp/sst119
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamania using guide RNA and Cas9. Nature Biotechnology, 31, 688–691. https://doi.org/10.1038/nbt.2654
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D. G., & Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamania using Cas9 RNA-guided endonuclease. Nature Biotechnology, 31, 691–693. https://doi.org/10.1038/nbt.2655
Vaia, G., Pavese, V., Moglia, A., Cristofori, V., & Silvestri, C. (2022). Knockout of phytoene desaturase gene using CRISPR/Cas9 in highbush blueberry. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2022.1074541
Donini, M., & Marusic, C. (2019). Current state-of-the-art in plant-based antibody production systems. Biotechnology Letters, 41, 335–346. https://doi.org/10.1007/s10529-019-02651-z
Kaur, M., Manchanda, P., Kalia, A., Ahmed, F. K., Nepovimona, E., Kuca, K., et al. (2021). Agroinfiltration mediated scalable transient gene expression in genome edited crop plants. International Journal of Molecular Sciences, 22(19), 10882. https://doi.org/10.3390/ijms221910882
Jia, H., Orbovic, V., Jones, J. B., & Wang, N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnology Journal, 14(5), 1291–1301. https://doi.org/10.1111/pbi.12495
Jia, H., Zhang, Y., Orbovic, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnology Journal, 15(7), 817–823. https://doi.org/10.1111/pbi.12677
Jia, H., Xu, J., Orbovic, V., Zhang, Y., & Wang, N. (2017). Editing citrus genome via SaCas9/sgRNA system. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2017.02135
Jia, H., Wang, Y., Su, H., Huang, X., & Wang, N. (2022). LbCas12-D156R efficiently edits LOB1 effector binding elements to generate canker-resistant citrus plants. Cells, 11(3), 315. https://doi.org/10.3390/cells11030315
Sendin, L. N., Filippone, M. P., Orce, I. G., Rigano, L., Enrique, R., & Pena, L. (2012). Transient expression of pepper Bs2 gene in Citrus limon as an approach to evaluate its utility for the management of citrus canker disease. Plant Pathology, 61(4), 648–657. https://doi.org/10.1111/j.1365-3059.2011.02558.x
Huang, X., Wang, Y., & Wang, N. (2022). Highly efficient generation of canker-resistant sweet orange enabled by an improved CRISPR/Cas9 System. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2021.769907
Jia, H., & Wang, N. (2014). Targeted Genome Editing of Sweet Orange Using Cas9/sgRNA. PLoS ONE, 9(4), e93806. https://doi.org/10.1371/journal.pone.0093806
Mahawar, M. K., Jalgaonkar, K., Bibwe, B., Bhushan, B., Meena, V. S., & Sonkar, R. K. (2020). Post-harvest processing and valorization of Kinnow mandarin (Citrus reticulata L.): a review. Journal of Food Science and Technology., 57(3), 799–815. https://doi.org/10.1007/s13197-019-04083-z
Manchanda, P., Kaur, H., Mankoo, R. K., Kaur, A., Kaur, J., Kaur, S., & Sidhu, G. S. (2022). Optimization of extraction of bioactive phenolics and their antioxidant potential from callus and leaf extracts of Citrus sinensis (L.) Osbeck, C. reticulata Blanco and C. maxima (Burm.) Merr. Journal of Food Measurement and Characterization. https://doi.org/10.1007/s11694-022-01695-6
Manchanda, P., Kaur, H., Mankoo, R. K., Kaur, J., Kaur, M., & Sidhu, G. S. (2023). Effect of solvent ratio, temperature and time on extraction of bioactive compounds and their antioxidant potential from callus, leaf and peel extracts of Citrus pseudolimon Taraka. Journal of Food Measurement and Characterization. https://doi.org/10.1007/s11694-023-02111-3
Safdar, M. N., Kausar, T., Jabbar, S., Mumtaz, A., Ahad, K., & Saddozai, A. A. (2017). Extraction and quantification of polyphenols from kinnow (Citrus reticulata L.) peel using ultrasound and maceration techniques. Journal of Food and Drug Analysis, 25(3), 488–500. https://doi.org/10.1016/j.jfda.2016.07.010
Magda, R., Awad, A., & Selim, K. (2008). Evaluation of mandarin and navel orange peels as natural sources of antioxidant in biscuits. Alexandria Journal of Food Science and Technology, 5(2), 75–82. https://doi.org/10.12816/AJFS.2008.19647
Navarro, C., Abelenda, J., Cruz-Oro, E., Cueller, C., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature, 478, 166–176. https://doi.org/10.1038/nature10431
Calderon-Perez, B., Ramrez-Pool, J. A., Nunez-Munoz, L. A., Vargas-Harnandez, B. Y., Camacho-Romero, A., Lara-Villamar, M., et al. (2022). Engineering macromolecular trafficking into the citrus vasculature. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2022.818046
Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution, 38, 3022–3027.
Yuan, W. (1990). Culture medium for Xanthomonas campestris pv. oryzae. Journal of Applied Bacteriology, 69, 798–805.
Doyle, J. (1991). DNA protocols for plants in Molecular Techniques. Berlin: Springer.
Lei, Y., Lu, L., Liu, H. Y., Li, S., Xing, F., & Chen, L. L. (2014). CRISPR-P A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Molecular Plant, 7(9), 1494–1496. https://doi.org/10.1093/mp/ssu044
Cohen, S. N., Chang, A. C., & Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: Genetic transformation of Escherichia coli by R-factor DNA. Proceedings of the National Academy of Sciences, 69(8), 2110–2114. https://doi.org/10.1073/pnas.69.8.2110
Holsters, M., de Waele, D., Depicker, A., Messens, E., van Montagu, M., & Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Molecular and General Genetics, 163(2), 181–187. https://doi.org/10.1007/bf00267408
Koschmieder, J., Fehling-Kaschek, M., Schaub, P., Ghisla, S., Brausemann, A., Timmer, J., & Beyer, P. (2017). Plant-type phytoene desaturase: Functional evaluation of structural implications. PLoS ONE, 12(11), e0187628. https://doi.org/10.1371/journal.pone.0187628
Hooghvorst, I., Lopez-Cristoffiani, C., & Nogues, S. (2019). Efficient knockout of phytoene destaurase gene using CRISPR/Cas9 in melon. Science and Reports, 9, 17077. https://doi.org/10.1038/s41598-019-53710-4
Odipio, J., Alicai, T., Ingelbredcht, I., Nusinow, D. A., Bart, R., & Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Frontiers in Plant Science. https://doi.org/10.3389/fpls.2017.01780
Komatsu, H., Abdellatif, I. M. Y., Yuan, S., Ono, M., Nonaka, S., Eruza, H., et al. (2020). Genome editing in PDS genes of tomatoes by non-selection method and of Nicotiana benthamiana by one single guide RNA to edit two orthologs. Plant Biotechnol., 37(2), 213–221. https://doi.org/10.5511/plantbiotechnology.20.0527b
Nezhdanona, A. V., Slugina, M. A., Kulakova, A. V., Kamionskaya, A. M., Kochieva, E. Z., & Shchennikova, A. V. (2023). Effect of mosaic knockout of phytoene desturase gene NtPDS on biosynthesis of carotenoids in Nicotiana tabacum L. Russian Journal of Plant Physiology, 70, 116. https://doi.org/10.1134/S1021443723601271
Ntui, V. O., Tripathi, J. N., & Tripathi, L. (2020). Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa app.). Current Plant Biology, 21, 100128. https://doi.org/10.1016/j.cpb.2019.100128
Siddappa, S., Sharma, N., Salaria, N., Thakur, K., Pathania, S., Singh, B., et al. (2023). CRISPR/Cas9-mediated editing of phytoene desturase (PDS) gene in an important staple crop, potato. Biotechnology, 13, 129. https://doi.org/10.1007/s13205-023-03543-w
Wan, L., Wang, Z., Zhang, X., Zeng, H., Ren, J., Zhang, N., et al. (2023). Optimized Agrobacterium-mediated transformation and application of developmental regulators improve regeneration efficiency in melons. Genes, 14(7), 1432. https://doi.org/10.3390/genes14071432
Jalan, N., Aritua, V., Kumar, D., Yu, F., Jones, J. B., Graham, J. H., et al. (2011). Comparative genomic analysis of Xanthomonas axonopodis cv. Citrumelo F1, which causes citrus bacterial spot disease, and related strains provides insights into virulence and host specificity. Journal of Bacteriology, 193, 6342–6357. https://doi.org/10.1128/jb.05777-11
Scott, S. E., Eric, M., Marc, M., & Patricia, Z. (1985). Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature, 318, 624–629. https://doi.org/10.1038/318624a0
Cangelosi, G. A., Ankenbauer, R. G., & Nester, E. W. (1990). Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proceedings of the National Academy of Sciences, 87, 6708–6712. https://doi.org/10.1073/pnas.87.17.6708
Zhang, J., Boone, L., Kocz, R., Zhang, C., Binns, A. N., & Lynn, D. G. (2000). At the maize/Agrobacterium interface: Natural factors limiting host transformation. Chemistry & Biology, 7, 611–621.
Lu, Y., & Zhu, J. K. (2016). Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Molecular Plant. https://doi.org/10.1016/j.molp.2016.11.013
Sony, S. K., Kaul, T., Motelb, K. F. A., Thangaraj, A., Bharti, J., Kaul, R., et al. (2023). CRISPR/Cas9-mediated homology donor repair base editing confers glyphosate resistance to rice (Oryza sativa L.). Frontiers in Plant Science. https://doi.org/10.3389/fpls.2023.1122926
Sun, Y., Zhang, X., Wu, C., He, Y., Ma, Y., Hou, H., et al. (2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Molecular Plant, 9, 628–631. https://doi.org/10.1016/j.molp.2016.01.001
Chen, X., Lu, X., Shu, N., Wang, S., Wang, J., Wang, D., et al. (2017). Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/CAs9 system. Science and Reports, 7, 44304. https://doi.org/10.1038/srep44304
Cai, Y., Chen, L., Zhang, Y., Yuan, S., Su, Q., Sun, S., et al. (2020). Target base pair editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnology Journal, 18, 1996–1998. https://doi.org/10.1111/pbi.13386
Zhao, H., Tan, Z., Wen, X., & Wang, Y. (2017). An improved syringe agroinfitration protocol to enhance transformation efficiency by combinative use of 5-azacytidine, ascorbic acid and Tween-20. Plants, 6(1), 9. https://doi.org/10.3390/plants6010009
Li, X., Ahlman, A., Yan, X., Lindgren, H., & Zhu, L. (2010). Genetic transformation of the oilseed crop Crambe abyssinica. Plant Cell Tissue Organ Culture., 100, 149–156. https://doi.org/10.1007/s11240-009-9630-y
Chevreau, E., Dousset, N., Joffrion, C., Richer, A., Charrier, A., & Vergne, E. (2019). Agroinfiltration is a key factor to improve the efficiency of apple and pear transformation. Scientia Horticulturae, 251, 150–154. https://doi.org/10.1016/j.scienta.2019.03.003
Zhang, Y., Chen, M., Siemiatkowska, B., Toleco, M. R., Jing, Y., Strotmann, V., et al. (2020). A highly efficient Agrobacterium-mediated method for transient gene expression and functional studies in multiple plant species. Plant Communications, 1, 100028. https://doi.org/10.1016/j.xplc.2020.100028
Acknowledgements
The authors thank Dr. Prashant Mohanpuria, School of Agricultural Biotechnology, Punjab Agricultural University, for providing pKO-119 plasmid procured from Tokushima University, Japan.
Funding
The work is supported by the Department of Biotechnology under the Centre of Excellence Project entitled, ‘Development and Integration of Advanced Genomic Technologies for Targeted Breeding’ CSS-27 (PC-6372).
Author information
Authors and Affiliations
Contributions
PM designed the experiment, arranged the plant material and laboratory facilities, guided FK in conducting the research work and wrote the manuscript. HK conducted data analysis and wrote the manuscript. GSS provided kinnow plants for carrying out Agroinfiltration experiments. MSH provided the Xcc inoculum for carrying out the experiment. PC edited the manuscript. NSB was the mentor of PM and provided the idea for transient genetic transformation in kinnow. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors report there are no conflict of interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Manchanda, P., Kaur, H., Khan, F. et al. Agroinfiltration-based transient genome editing for targeting phytoene desaturase gene in kinnow mandarin (C. reticulata Blanco). Mol Biotechnol (2023). https://doi.org/10.1007/s12033-023-00980-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-00980-z