Abstract
An insect–plant interaction induced gall formation is where gall wasps change the plant development towards formation of new units to shield and nourish the evolving larvae. The targets of the insect signals and the mechanism of gall development are unknown. To show the molecular pathways that are responsive to the gall wasp, the proteomic approach was used to compare the gall with non-gall plant tissues. We studied three oak gall species (Cynips quercusfolii, Cynips longiventris, and Neuroterus quercusbaccarum) and the host plant (Quercus robur). Among the 21 identified proteins, 18 increased and three decreased in abundance in gall tissue, in comparison to the leaf tissues. Ten proteins were C. quercusfolii responsive, two only with this gall inducer, while seven increased in abundance. Eleven proteins were C. longiventris responsive, and two only with this gall inducer. Sixteen proteins were associated with gall formation by the N. quercusbaccarum and, in this, eight only with this gall inducer. A similar effect on protein abundance occurred as galls in leaf veins (for five proteins). For leaf blades, such a relation was not found. The role of each protein is discussed according to its involvement in the gall formation. Moreover, S-adenosyl methionine synthase, flavone 3-hydroxylase, stress- and pathogenesis-related proteins, and gamma carbonic anhydrase are associated with developmental regulation of plant tissue into a gall.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant gall induction is common in many insect families, but most of the species belong to only two groups: gall wasps and gall midges (Cook and Gullan 2004). The insect controls plant morphogenesis, inducing the formation of a new structure as a gall (Harper et al. 2004). There are three well-distinguished phases of gall formation: first initiation, subsequent growth, and final maturation (Rohfritsch and Shorthouse 1982; Rey 1992). Oviposition by the female wasp or midge onto the meristematic tissue starts the initiation phase. The insect controls the place of gall emergency (Giertych et al. 2013). The host usually reacts with necrosis of cells under the egg; the cells below divide and finally enclose the egg (Rey 1992; Brooks and Shorthouse 1998; Pilichowski and Giertych 2017). The gall chamber is formed through cell differentiation and is lined inside with inner-gall tissue, and the outer gall is built with cortical parenchyma. The growth phase primarily is composed of cell elongation, leading to gall expansion. The cell layers of the inner-gall tissue decrease during development because the larvae feed on them. The parenchyma cells are changed into nutritive cells as the larvae feed, and they produce lipids and proteins as well as amylases, which are involved in starch decomposition (Bronner 1992; Rey 1992; Brooks and Shorthouse 1998). During maturation, cell division stops, the galls lignify, the larvae mature and pupate, and finally, the gall tissue desiccates (Harper et al. 2004).
Cynipid gall generation is a result of the complicated interaction between the insect and the host plant. The larvae interact with the plant to switch normal plant development to ensure shelter, nutrients, and protection for their defence inside a gall (Stone et al. 2002). The template of the internal gall tissue is generally similar for all cynipid galls, with two distinguished layers, an outer cortical parenchyma and an inner gall, in which one (monolocular) or many (multilocular) larvae are housed (Harper et al. 2004). The inner-gall chamber is surrounded by a hard layer of sclerenchyma (Guzicka et al. 2017), and its tissue is built of two kinds of cells: inner nutritive cells and nutritive parenchyma cells (Harper et al. 2004). During gall formation, many physiological, biochemical, and molecular changes occur in the tissue, showing meaningful distinction from the host-plant non-gall tissue (Harper et al. 2004). The signal pathway from the gall wasp to the host plant is almost unknown; however, gall formation requires continuous activity of unknown stimuli (Stone et al. 2002). Insect-herbivore induction causes anatomical and biochemical modification of the host-plant tissues into the gall. The gall inducers can manipulate the plant metabolism according to their own benefit (Detoni et al. 2010). Detoni et al. (2011) hypothesised that specific proteins, likely representing different metabolic classes, are associated with the physiological processes determining the host resistance to the gall-insect attack.
Although the number of publications about the manipulation of plant tissues by insects forming galls is large, how they reprogram the host-plant development is not still well characterised (Cornell 1983; Hartley and Lawton 1992; Schönrogge et al. 1998; Hartley 1998). To recognise such a mechanism would be of great interest for insect–plant interaction studies.
To identify the molecular mechanism involved in gall formation, we investigated changes in the oak gall proteomes induced by three cynipid wasps commonly found in this region: Cynips quercusfolii, Cynips longiventris, and Neuroterus quercusbaccarum. Analysis of the differences and similarities between Quercus robur gall tissues and non-gall tissues allows us to show the level of control exercised by the insect on the plant development, and the relationship between the gall and plant tissues. We assumed a high similarity of protein profiles for galls of each wasp species and a greater affinity of galls with leaf vein tissue than with leaf blade. Results are discussed in relation to system biology approaches (Staszak and Pawłowski 2012).
Methods and materials
Plant materials and experimental design
The samples of leaves and all gall species were collected from six (treated as a repetition) trees of English oak (Q. robur L.) growing at the edge of a forest in Tulce (Kobylepole Forest, Babki Forest District, Poland, 52°21′03″N; 17°04′16″E) in August 2012. This period is optimal because galls have reached the maximum size and maturity, but larvae are very small yet. Agamic generation galls induced by C. quercusfolii Linnaeus 1758, C, longiventris Linnaeus 1758, and N. quercusbaccarum Linnaeus 1758, and leaves, which were divided into veins and laminas, were collected from every tree. An analytical sample for each tree was collected (for each insect species) from a few to several galls, depending on the size of each, and several leaves were divided into laminas and veins. A total of 30 samples were analysed. The gall tissue, leaf veins, and laminas were dissected 1 h after harvest and frozen in liquid nitrogen and stored at −70 °C.
Protein extraction
Extracts were prepared from three types of galls (without the insects), leaf blades, and leaf veins. At least three independent protein samples were prepared, as already presented (Faurobert et al. 2007; Pawłowski 2009; Staszak and Pawłowski 2014). Ground material was resuspended in three volumes of extraction buffer (consisting of 700 mM saccharose, 500 mM tris, pH 8, 100 mM KCl, 2% (v/v) β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride, pH 8.5) and kept for 10 min on ice. Subsequently, tris-saturated phenol was added in equal volume. Samples were stirred for 10 min and centrifuged (10 min, 5525g, 4 °C) to isolate the phenolic and aqueous phases. The phenolic phase was extracted again with the same volume of extraction buffer. Subsequently, centrifugation was repeated, and five volumes of precipitation solution (0.1 M ammonium acetate in methanol) were added to the recovered phenol phase. Proteins were precipitated at −20 °C overnight. After centrifugation for 10 min (5525g, 4 °C), the protein pellet was washed three times with the precipitation solution and once with acetone. Each washing step was followed by 5 min of centrifugation as described above. After drying under a vacuum, the pellet was resuspended in lysis buffer [9 M urea, 4% (w/v) CHAPS, 0.5% (v/v) Triton X-100, 20 mM dithiothreitol (DTT), and 1.2% (v/v) pharmalyte, pH 3–10] and the protein concentration was measured according to a modified Bradford assay (Ramagli and Rodriguez 1985).
Protein electrophoresis, 2-DE IEF/SDS-PAGE
All analyses were conducted at 25 °C with at least three biological replicas. Proteins (700 mg for colloidal Coomassie blue) were first separated according to their charge on rehydrated Immobiline dry strips (24 cm, containing linear gradient of pH 4–7) with the rehydration buffer [6 M urea, 2 M thiourea, 2% (w/v) CHAPS, 20 mM (w/v) DTT, and 0.5% (v/v) pharmalyte, pH 4–7] on Ettan IPGphor 3 IEF System (GE Healthcare Life Science). The programme for isoelectric focusing was according to the manufacturer’s suggestions for 24-cm strips. The strips were either stored at −80 °C or they were directly treated for 10 min with equilibration solution I [6 M urea, 1.5 M Tris–HCl, pH 8.8, 30% (v/v) glycerol, 2% (w/v) SDS, and 1% (w/v) DTT] and for the same time with equilibration Solution II [Solution I supplemented with 2.5% (w/v) iodoacetamide, without DTT] and subjected for the second dimension (SDS-PAGE) run.
For SDS-PAGE, precasted Ettan DALT 12.5% (w/v) polyacrylamide gels (GE Healthcare) and the Ettan Dalt Six electrophoresis unit were used. Conditions for the run were as follows: 1 h at 80 V and 5 h at 500 V. Molecular weight markers (GE Healthcare) were applied. After electrophoresis, proteins were stained with colloidal Coomassie blue, which allowed visualisation, quantification, and mass spectrometry (MS) analysis (Neuhoff et al. 1988).
Proteome analysis
The protein patterns were analysed after scanning using the 2D Image Master 7 Platinum program (GE Healthcare). The two-dimensional (2D) patterns (three from three independent biological samples) were aligned and matched after spot detection, and the normalised spot volumes were ascertained quantitatively. The per cent volume was calculated for each matched spot, as the volume divided by the total volume of matched spots. Abundantly variable spots (significantly changed between gel sets/classes, i.e., central tendency, dispersion, and overlapping measures were used) underwent analysis of variance (ANOVA) and the Tukey–Kramer HSD test (JMP software, SAS Institute, Cary, USA) to choose spots that significantly varied (p < 0.05) in abundance for the five factors: the gall types and leaf parts. Protein spots that exhibited at least a 1.5-fold increase or decrease in their relative abundance between galls and leaf tissues were considered differentially expressed. Fold change was calculated as log2 (Meunier et al. 2005). The significant alternating proteins were identified by MS.
Mass spectrometry (MS)
The protein spots underwent a standard ‘in-gel digestion’ processing; they were reduced with 10 mM (w/v) DTT (for 30 min at 56 °C), alkylated 45 min with 55 mM iodoacetamide (in the dark at room temperature), and digested overnight with trypsin (Promega, Madison, WI, USA) in 25 mM ammonium bicarbonate (25 ng/µl). The resulting peptides were eluted from the polyacrylamide with 0.1% (v/v) TFA in 2% (v/v) ACN.
Peptides were analysed by liquid chromatography and the mass spectrometer device at the Institute of Biochemistry and Biophysics, the Laboratory of Mass Spectrometry at the Polish Academy of Sciences, Warsaw, Poland. Samples were concentrated and desalted on an RP-C18 pre-column (nanoACQUITY Symmetry® C18, Waters, Milford, MA, USA), and peptides were separated on a nano-ultra performance liquid chromatography (UPLC) RP-C18 column (Waters, BEH130 C18 column, 75 µm id, 250 mm long) of a nanoACQUITY UPLC system, using a linear acetonitrile gradient [0–60% (v/v) ACN for 120 min] in the presence of 0.05% (v/v) formic acid with a flow rate of 150 nl/min. The column outlet was directly coupled to the electrospray ionisation (ESI) ion source of the orbitrap velos type mass spectrometer (Thermo Electron Corp., San Jose, CA, USA), working in the regime of data dependent MS to MS/MS switch. An electrospray voltage of 1.5 kV was used. A blank run preventing cross-contamination from previous samples preceded each analysis.
The mascot search algorithm (http://www.matrixscience.com) was used for protein identification, against the NCBInr (http://www.ncbi.nig.gov) databases. The mascot search probability-based molecular weight search (MOWSE) score was used. The ion score was −10 × log(P), where P was the probability that the observed match was a random event. Peptides with a mascot score exceeding the threshold value, corresponding to a <5% false positive rate, as calculated by the mascot procedure, were considered to be positively identified.
Identified proteins were functionally grouped according to the KEGG database (http://www.genome.jp/kegg/pathway.html) (Kanehisa et al. 2012), and a Venn diagram of protein quantification was prepared according to bioinformatics and evolutionary genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Results
Protein patterns from three gall types in leaf blades and veins were analysed to estimate proteins involved in the gall-formation process. Proteome maps were established, which displayed on, average, 359 Coomassie blue-stained spots. A total set of 21 spots (6% of the total spots) showing significant changes in volume (ANOVA, the Tukey–Kramer HSD test, and fold change >1.5) were identified by MS. Identified proteins displayed significant differential abundance for the gall type and the part of the leaf.
The gels from each variant were used to create master gels, showing the results of statistical analysis (Fig. 1a–e). Among the 21 identified proteins (Table 1; Fig. 2), 18 increased and three decreased in abundance in the gall tissue, compared to the leaf tissues. Ten proteins were C. quercusfolii responsive, two only with this gall inducer, while seven increased in abundance. Eleven proteins were C. longiventris responsive, two only with this gall inducer, while eight increased in abundance. Sixteen proteins were associated with gall formation by N. quercusbaccarum, in this, eight only with this gall inducer, while 13 increased in abundance. For five proteins, the influence of the gall wasp was statistically the same and was simultaneously significantly different from the whole leaf (three proteins) or leaf blade (two proteins). A similar effect on protein abundance, as galls had leaf vein (for five proteins). For leaf blades, such a relation was not observed.
Positions of 21 proteins that varied in abundance on 2D colloidal blue-stained gels of Quercus robur galls initiated by Cynips quercusfolii (a), Cynips longiventris (b), Neuroterus quercusbaccarum (c), and in control (without galls) of Q. robur leaf blades (d) and veins (e). Labels show the proteins are abundantly variable between galls and leaf tissues. The numbers of these 21 identified proteins present in the gels correspond to those listed in Table 1
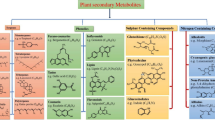
The identified proteins were further grouped, based on their functional categories, into four classes: metabolism, genetic information processing, cellular processes, and unclassified (Table 1; Fig. 3).
Classification of the 21 identified abundantly variable proteins to functional categories according to the KEGG database. The percentage of proteins in each class based on the number of all the identified proteins is shown as an indication of how many were upregulated (dark portion of each bar) or downregulated (white portion of each bar). Proteins associated with gall formation by Cynips quercusfolii (a), C. longiventris (b), and Neuroterus quercusbaccarum (c)
Discussion
The proteomic approach is broadly used for the investigation of many developmental processes in plants (e.g., Pawłowski 2009, 2010; Pawłowski and Staszak 2016). There are just a few research studies carried out on the molecular mechanism, especially at the proteomic level, associated with gall formation. The protein analysis of inner-gall and plant tissue showed the distinct protein profiles and identified some inner-gall proteins (Schönrogge et al. 2000). These proteins, which are characteristic only for gall tissues, may enable understanding of how the larvae change the plant development to form galls. The NAD-dependent formate dehydrogenase (FDH) was identified in the inner-gall tissue in two species. The FDHs are a nuclear-encoded proteins found in the mitochondria and expressed under stress (Hourton-Cabassa et al. 1998). Schönrogge et al. (2000) also detected a biotin carboxyl carrier protein (BCCP), a part of acetyl CoA-carboxylase (ACCase), associated with the synthesis of triacylglycerol lipids, an energy supplier (Elborough et al. 1996). The upregulation of BCCP demonstrates an increase in lipid synthesis to provide sufficient nutrients for the growing larvae. Schönrogge et al. (2000) suggested that identified proteins are associated with respiratory stress arising in galls, and seed-specific proteins are involved in gall formation.
The insect gut participates in diverse physiological processes, for example in host–insect interactions and regulations of host development (Nation 2002). The most impressive characteristic of the gall midge gut larval transcriptome is the presence of a large quantity of small secretory protein (SSP) transcripts (Zhang et al. 2010). The SSPs likely act in plant–insect interactions and might be valid regulators of gall development (Zhang et al. 2010). A gall insect can restrain plant growth, reprogram plant gene transcription, and stimulate nutritive tissues (Anderson and Harris 2006; Harris et al. 2006; Liu et al. 2007).
Our results showed that the protein patterns differ between galls and leaves. We observed protein characteristics for Cynips galls and for N. quercusbaccarum. Proteomes of Cynips galls were much more similar to each other than to N. quercusbaccarum. All that suggests different influences of gall wasps from different species on the host plant. The function of the investigated proteins and the associated metabolic pathways according to involvement in gall formation will be discussed in subsequent sections. The discussion will follow the magnitude of fold-change values (Table 2).
Metabolism
The S-adenosyl methionine (SAM) synthase belongs to the secondary metabolism class (spots 53, 128, and 129 increased and spot 54 decreased in abundance in the galls). The SAM synthase catalyses the formation of the SAM from methionine and ATP. The SAM is a substrate in the nicotianamine, ethylene, and polyamine biosynthesis reactions and during plant growth and development, ensures the methyl groups for the most methylation reactions (Ravanel et al. 1998; Sauter et al. 2013). Ethylene controls many features of plant growth and development from seed germination through the root and stem formation to fruit ripening and is a key mediator of stress responses in plants (Merchante et al. 2013). It controls plant development and growth by regulating the meristem activity and cell-wall structure (Vandenbussche et al. 2012). Among these functions of ethylene is the regulation of ‘Rhizobium’-induced nodulation in legumes (Gresshoff et al. 2009), a process that is, in some way, similar to gall formation. Polyamines actively participate in cell division, cell specification, and transcriptional, translational, and post-translational regulation of genes, transcripts, and proteins (Tiburcio et al. 2014). In plants, they are involved in developmental processes, such as organogenesis, floral and fruit expansion, and response to the influence of abiotic and biotic stress factors (Tiburcio et al. 2014). Interestingly, biotin synthase (spot 253, found only in N. quercusbaccarum) is also a SAM radical enzyme that is involved in biotin metabolism. Biotin (essential vitamin H/B8) is an enzyme cofactor implicated in reactions of carboxylation and decarboxylation of fatty acid and carbohydrate metabolism (Fugate and Jarrett 2012). The biotinylation of these reaction enzymes is a post-translational modification, allowing the transformation of inactive proteins into their active forms (Alban 2011). Summarising, the SAM metabolism, being a central point in nitrogen metabolism and other metabolites, can play an essential role in gall formation by influencing gene expression pathways, cell division, and tissue differentiation.
Flavanone 3-hydroxylase (spot 187), which arose in galls, is a main enzyme in the flavonoid biosynthesis (Winkel-Shirley 2001). A main physiological function of the flavonoid pigments is the lure of flower pollinators and seed dispersers. This aim is achieved by beautiful pigmentation of flowers, fruits, etc. Besides that, flavonoids also play a pivotal function in plant and microbe signalling and in stress protection, mainly against excessive UVB radiation (Ballaré 2014). Their role in galls can be associated with the colour of the gall surface, which may play a signalling role between the plant and insect. Differential flavonoid accumulation and flavanone 3-hydroxylase expression was observed in legume plant nodules, in comparison with root tissue (Charrier et al. 1998).
Catalase (spot 125) is a ubiquitous enzyme involved in the cell protection from the toxic effect of peroxides (Sofo et al. 2015). Enzyme catalyses the transition of hydrogen peroxide to water and molecular oxygen. Catalases also utilise hydrogen peroxide to oxidise various substrates, such as alcohol or phenols. Catalase arose in galls and reached the highest abundance in C. quercusfolii galls in comparison to those of leaf tissues. A similar effect was observed in the galls of Alstonia scholaris caused by Pseudophacopteron alstonium (Yang et al. 2015). According to these authors and our results, it can be concluded that an increase in catalase activity is associated with the defensive response of the host-plant against insect activity.
The ATP synthase (spots 9 and 63, increased in galls) provides energy for the growing gall tissue. Enolase is also associated with energy provision (spots 58 and 134, increased in galls) and is an enzyme involved in glycolysis, a process in which ATP is synthesised.
The UDP-glucose pyrophosphorylase (UGPase, spots 61 and 62) is a key enzyme implicated in the synthesis of UDP-glucose, which is involved in glycosylation reactions (Decker et al. 2012). These include synthesis of sucrose, polysaccharides, glycoproteins, glycolipids, sulpholipids, and secondary metabolites. The UDP sugars are, by far, the major substrates for polysaccharide synthesis (Kotake et al. 2010). In the present study, a decrease in UGPase abundance was indicated in gall tissues in comparison to leaf tissues (Table 2). The role of UGPase in gall formation is difficult to distinguish; however, it can be associated with the synthesis of nucleotide sugars, which serve as glycosyl donors or with starch synthesis. Upregulation of the UGPase gene in Populus deltoids leaves resulted in reduced sugar and starch levels (Payyavula et al. 2014). Starch is a main part of the nutritive layer in the gall. This suggests that starch synthesis can also increase in galls, providing nutrition for the larvae.
Aldehyde dehydrogenase 2 (ALDH, spot 66, increased in C. longiventris) catalyses the reactive aldehyde oxidation into their adequate carboxylic acids (Kirch et al. 2005). The ALDHs use either NAD+ or NADP+ as a cofactor to convert aldehydes to carboxylic acids plus NADH or NADPH. Compounds with aldehydic functional groups are generated as important intermediates in many catabolic and biosynthetic pathways of carbohydrates, lipids, vitamins, steroids, and amino acids. Moreover, the ALDH-mediated generation of NADH/NADPH represents a major source of reducing equivalents required for maintaining cellular redox balance (Brocker et al. 2012). In addition, ALDHs act as ‘aldehyde scavengers’ during lipid peroxidation where they remove reactive aldehydes entailed by the oxidative degradation of lipid membranes (Brocker et al. 2012). ALDHs that have been characterised to date, the most have been involved in different processes and have putative substantial roles in growth and development of the plant (Tian et al. 2015). For example, the rice ALDH7 is essential for seed maturation and longevity (Shin et al. 2009). The ALDH2 family members in plants detoxify acetaldehyde to acetate, which can be later used in fatty acid synthesis (Wei et al. 2009). The ALDH2 role in gall development can be associated with synthesis of nutrition compounds used by gall wasp larvae.
Chalcone synthase (CHS, spot 189) synthetises polyketides, a group of diverse secondary metabolites as anthocyanins and flavonoids in plants. Plants have evolutionarily recruited the CHSs to adapt to the changing environment, especially to biotic stresses (Han et al. 2014). The expression of CHS was significantly upregulated during gall formation in fig flowers (Martinson et al. 2015). A similar effect, an increase in abundance, was indicated in investigated galls, in comparison to leaf blades. The flavonoids play diverse roles in plant–insect interactions (Simmonds 2001, 2003). They have been associated putatively with host recognition and insect reception (Martinson et al. 2015). However, they can diminish the growth and survivorship of insects (Abou-Zaid et al. 1993). Flavonoids can also stimulate oviposition of Lepidoptera (Honda 1990). The function CHS can play in galls is probably associated with the stimulation of oviposition by the next larvae, a hypothesis that was also postulated by Martinson et al. (2015).
Genetic information processing
Cytosolic Class II low molecular weight HSP (spot 114) and HSP 17.4 (spot 115) are small stress-induced molecular chaperones belonging to the HSP20/sHSP family. Their role is associated with protein processing in the endoplasmic reticulum, either by direct interaction with or by affecting signalling components involved in the initiation of the hypersensitive response (Sarkar et al. 2009). Recent data have shown that they are implicated in the response to biotic stresses (Lopes-Caitar et al. 2013). In the present study, they reached the highest significant abundance in N. quercusbaccarum. They are probably implicated in the response to stress associated with insect activity.
Cellular processes
Actin (spot 45) is a base protein of the cytoskeleton participating in a large number of essential cellular processes, including cell division, cell elongation, cell signalling, vesicle and organelle movement, and the creation and preservation of cell connections and shape (Volkmann and Baluška 1999; Thomas et al. 2009; Higaki et al. 2010). Increase in actin accumulation during gall formation can be associated with cell division in the developing new tissues.
Unclassified
Protein DDB_G0288155 (spot 23, increased in galls) exhibits high sequence homology to gamma CA proteins. The CAs catalyse the synthesis of bicarbonate (and protons) from carbon dioxide and water (Park et al. 2012). Gamma CAs were discovered in mitochondrial Complex I, where they are responsible for the carbon transportation between mitochondria and chloroplasts that enhance the performance of carbon dioxide fixation (Ferry 2010). Their role in gall formation can be associated with the higher demand for the carbon skeleton during development.
Stress and pathogenesis-related proteins (spot 98, increased in N. quercusbaccarum) are a homolog of pathogenesis-related class 10 (PR-10) proteins, which are produced as a defence response to various pathogens (Pasternak et al. 2006). It also has a similar sequence to PYR/PYL/RCAR proteins, which are receptors of ABA (Klingler et al. 2010). Its role in the ABA transduction pathway can be proof for the involvement of ABA in regulating cynipid gall formation. On one hand, stress and pathogenesis-related proteins can play a protective role against insects; on the other hand, by involvement in ABA signalling, it may be responsive for initiation/building of the isolation structure (gall) from the plant tissues. Liu et al. (2010) showed that reactive oxygen species are implicated in plant defence against a gall midge. In rice class III, peroxidases play a role in the response against Hessian fly attacks. That can also be an explanation for the role of stress and pathogenesis-related proteins in controlling gall formation.
Author contribution statement
Conceived and designed the experiments: MJG, TAP. Performed the experiments: AMS, TAP. Analysed the data: TAP. Contributed reagents/materials/analysis tools: MJG, PK, TAP. Wrote the paper: TAP. Critically revised the paper: MJG, PK, AMS. All authors read and approved the manuscript.
References
Abou-Zaid MM, Beninger CW, Arnason JT, Nozzolillo C (1993) The effect of one flavone, two catechins and four flavonols on mortality and growth of the European corn borer (Ostrinia nubilalis Hubner). Biochem Syst Ecol 21:415–420. doi:10.1016/0305-1978(93)90099-D
Alban C (2011) Chapter 2—Biotin (vitamin B8) synthesis in plants. In: Rebeille F (ed) Advances in botanical research. Academic Press, London, pp 39–66
Anderson KG, Harris MO (2006) Does R gene resistance allow wheat to prevent plant growth effects associated with hessian fly (Diptera: Cecidomyiidae) attack? J Econ Entomol 99:1842–1853. doi:10.1093/jee/99.5.1842
Ballaré CL (2014) Light regulation of plant defense. Ann Rev Plant Biol 65(1):335–363. doi:10.1146/annurev-arplant-050213-040145
Brocker C, Vasiliou M, Carpenter S et al (2012) Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237:189–210. doi:10.1007/s00425-012-1749-0
Bronner R (1992) The role of nutritive cells in the nutrition of cynipids and cecidomyiids. In: Shorthouse JD, Rohfritsch O (eds) Biology of insect-induced galls. Oxford University Press, New York, pp 118–140
Brooks SE, Shorthouse JD (1998) Developmental morphology of stem galls of Diplolepis nodulosa (Hymenoptera: Cynipidae) and those modified by the inquiline Periclistus pirata (Hymenoptera: Cynipidae) on Rosa blanda (Rosaceae). Can J Bot 76:365–381. doi:10.1139/b98-001
Charrier B, Trinh H, Poirier S et al (1998) Flavanone 3-hydroxylase (F3H) expression and flavonoid localization in nodules of three legume plants reveal distinct tissue specificities. Mol Plant Microbe Interact 11:924–932. doi:10.1094/MPMI.1998.11.9.924
Cook LG, Gullan PJ (2004) The gall-inducing habit has evolved multiple times among the eriococcid scale insects (Sternorrhyncha: Coccoidea: Eriococcidae). Biol J Linn Soc 83:441–452. doi:10.1111/j.1095-8312.2004.00396.x
Cornell HV (1983) The secondary chemistry and complex morphology of galls formed by the Cynipinae (Hymenoptera): why and how? Am Midl Nat 110:225. doi:10.2307/2425263
de Detoni ML, Vasconcelos EG, Scio E et al (2010) Differential biochemical responses of Calliandra brevipes (Fabaceae, Mimosoidae) to galling behaviour by Tanaostigmodes ringueleti and T. mecanga (Hymenoptera, Tanaostigmatidae). Aust J Bot 58:280–285. doi:10.1071/BT09213
de Detoni ML, Vasconcelos EG, Ribeiro Gomes Maia AC et al (2011) Protein content and electrophoretic profile of insect galls on susceptible and resistant host plants of Bauhinia brevipes Vogel (Fabaceae). Aust J Bot 59:509–514. doi:10.1071/BT11104
Decker D, Meng M, Gornicka A et al (2012) Substrate kinetics and substrate effects on the quaternary structure of barley UDP-glucose pyrophosphorylase. Phytochemistry 79:39–45. doi:10.1016/j.phytochem.2012.04.002
Elborough KM, Winz R, Deka RK et al (1996) Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit farm of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem J 315:103–112
Faurobert M, Mihr C, Bertin N et al (2007) Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol 143:1327–1346. doi:10.1104/pp.106.092817
Ferry JG (2010) The γ class of carbonic anhydrases. Biochim Biophys Acta BBA—Proteins Proteom 1804:374–381. doi:10.1016/j.bbapap.2009.08.026
Fugate CJ, Jarrett JT (2012) Biotin synthase: insights into radical-mediated carbon–sulfur bond formation. Biochim Biophys Acta BBA—Proteins Proteom 1824:1213–1222. doi:10.1016/j.bbapap.2012.01.010
Giertych MJ, Jagodzinski AM, Karolewski P (2013) Spatial distribution of Cynips quercusfolii (Hymenoptera: Cynipidae) galls on leaves and within the crowns of oak trees. Eur J Entomol 110:657–661
Gresshoff PM, Lohar D, Chan P-K et al (2009) Genetic analysis of ethylene regulation of legume nodulation. Plant Signal Behav 4:818–823. doi:10.4161/psb.4.9.9395
Guzicka M, Karolewski P, Giertych MJ (2017) Structural modification of Quercus petraea leaf caused by Cynips quercusfolii—histological study of galls. J Plant Interact 12:7–13. doi:10.1080/17429145.2016.1269214
Han Y, Zhao W, Wang Z et al (2014) Molecular evolution and sequence divergence of plant chalcone synthase and chalcone synthase-like genes. Genetica 142:215–225. doi:10.1007/s10709-014-9768-3
Harper LJ, Schonrogge K, Lim KY et al (2004) Cynipid galls: insect-induced modifications of plant development create novel plant organs. Plant Cell Environ 27:327–335. doi:10.1046/j.1365-3040.2004.01145.x
Harris KM, Sato S, Uechi N, Yukawa J (2006) Redefinition of Oligotrophus (Diptera: Cecidomyiidae) based on morphological and molecular attributes of species from galls on Juniperus (Cupressaceae) in Britain and Japan. Entomol Sci 9:411–421. doi:10.1111/j.1479-8298.2006.00187.x
Hartley SE (1998) The chemical composition of plant galls: are levels of nutrients and secondary compounds controlled by the gall-former? Oecologia 113:492–501. doi:10.1007/s004420050401
Hartley SE, Lawton JH (1992) Host-plant manipulation by gall-insects: a test of the nutrition hypothesis. J Anim Ecol 61:113. doi:10.2307/5514
Higaki T, Kojo KH, Hasezawa S (2010) Critical role of actin bundling in plant cell morphogenesis. Plant Signal Behav 5:484–488. doi:10.4161/psb.10947
Honda K (1990) Identification of host-plant chemicals stimulating oviposition by swallowtail butterfly, Papilio protenor. J Chem Ecol 16:325–337. doi:10.1007/BF01021768
Hourton-Cabassa C, Ambard-Bretteville F, Moreau F et al (1998) Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol 116:627–635. doi:10.1104/pp.116.2.627
Kanehisa M, Goto S, Sato Y et al (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114. doi:10.1093/nar/gkr988
Kirch H-H, Schlingensiepen S, Kotchoni S et al (2005) Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol 57:315–332. doi:10.1007/s11103-004-7796-6
Klingler JP, Batelli G, Zhu J-K (2010) ABA receptors: the START of a new paradigm in phytohormone signalling. J Exp Bot 61:3199–3210. doi:10.1093/jxb/erq151
Kotake T, Hirosawa C, Ando Y, Tsumuraya Y (2010) Generation of nucleotide sugars for biomass formation in plants. Plant Biotechnol 27:231–236. doi:10.5511/plantbiotechnology.27.231
Liu X, Bai J, Huang L et al (2007) Gene expression of different wheat genotypes during attack by virulent and avirulent Hessian fly (Mayetiola destructor) larvae. J Chem Ecol 33:2171–2194. doi:10.1007/s10886-007-9382-2
Liu X, Williams CE, Nemacheck JA et al (2010) Reactive oxygen species are involved in plant defense against a gall midge. Plant Physiol 152:985–999. doi:10.1104/pp.109.150656
Lopes-Caitar VS, de Carvalho MC, Darben LM et al (2013) Genome-wide analysis of the Hsp 20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom 14:577. doi:10.1186/1471-2164-14-577
Martinson EO, Hackett JD, Machado CA, Arnold AE (2015) Metatranscriptome analysis of fig flowers provides insights into potential mechanisms for mutualism stability and gall induction. PLoS One 10:e0130745. doi:10.1371/journal.pone.0130745
Merchante C, Alonso JM, Stepanova AN (2013) Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol 16:554–560. doi:10.1016/j.pbi.2013.08.001
Meunier B, Bouley J, Piec I et al (2005) Data analysis methods for detection of differential protein expression in two-dimensional gel electrophoresis. Anal Biochem 340:226–230. doi:10.1016/j.ab.2005.02.028
Nation JI (2002) Digestion. In: Nation JI (ed) Insect physiology and biochemistry. CRC Press, Boca Raton, pp 27–64
Neuhoff V, Arold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric-focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262. doi:10.1002/elps.1150090603
Park H-M, Park J-H, Choi J-W et al (2012) Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch. Acta Crystallogr Sect Biol Crystallogr 68:920–926. doi:10.1107/S0907444912017210
Pasternak O, Bujacz GD, Fujimoto Y et al (2006) Crystal structure of Vigna radiata cytokinin-specific binding protein in complex with zeatin. Plant Cell 18:2622–2634. doi:10.1105/tpc.105.037119
Pawłowski TA (2009) Proteome analysis of Norway maple (Acer platanoides L.) seeds dormancy breaking and germination: influence of abscisic and gibberellic acids. BMC Plant Biol 9:48. doi:10.1186/1471-2229-9-48
Pawłowski TA (2010) Proteomic approach to analyze dormancy breaking of tree seeds. Plant Mol Biol 73:15–25. doi:10.1007/s11103-010-9623-6
Pawłowski TA, Staszak AM (2016) Analysis of the embryo proteome of sycamore (Acer pseudoplatanus L.) seeds reveals a distinct class of proteins regulating dormancy release. J Plant Physiol 195:9–22. doi:10.1016/j.jplph.2016.02.017
Payyavula RS, Tschaplinski TJ, Jawdy SS et al (2014) Metabolic profiling reveals altered sugar and secondary metabolism in response to UGPase overexpression in Populus. BMC Plant Biol 14:265. doi:10.1186/s12870-014-0265-8
Pilichowski S, Giertych MJ (2017) Gall abundance and leaf size as factors affecting the hypersensitive reaction in the common beech (Fagus sylvatica). Baltic For 23(2) (in press)
Ramagli LS, Rodriguez LV (1985) Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 6:559–563. doi:10.1002/elps.1150061109
Ravanel S, Gakière B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci 95:7805–7812
Rey LA (1992) Developmental morphology of two types of hymenopterous galls. In: Shorthouse JD, Rohfritsch O (eds) Biology of insect-induced galls. Oxford University Press, New York, pp 87–101
Rohfritsch O, Shorthouse JD (1982) Insect galls. Molecular biology of plant tumors. Academic Press, New York
Sarkar NK, Kim Y-K, Grover A (2009) Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genom 10:393. doi:10.1186/1471-2164-10-393
Sauter M, Moffatt B, Saechao MC et al (2013) Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451:145–154. doi:10.1042/BJ20121744
Schönrogge K, Harper LJ, Brooks SE et al (1998) Reprogramming plant development: two approaches to study the molecular mechanism of gall formation. In: Csoka G, Mattson WJ, Stone GN, Price PW (eds) Biology of gall-inducing arthropods. United States Department of Agriculture Forest Service, North Central Research Station, St Paul, pp 153–160
Schönrogge K, Harper LJ, Lichtenstein CP (2000) The protein content of tissues in cynipid galls (Hymenoptera: Cynipidae): similarities between cynipid galls and seeds. Plant Cell Environ 23:215–222. doi:10.1046/j.1365-3040.2000.00543.x
Shin J-H, Kim S-R, An G (2009) Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiol 149:905–915. doi:10.1104/pp.108.130716
Simmonds MSJ (2001) Importance of flavonoids in insect–plant interactions: feeding and oviposition. Phytochemistry 56:245–252. doi:10.1016/S0031-9422(00)00453-2
Simmonds MSJ (2003) Flavonoid–insect interactions: recent advances in our knowledge. Phytochemistry 64:21–30. doi:10.1016/S0031-9422(03)00293-0
Sofo A, Scopa A, Nuzzaci M, Vitti A (2015) Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int J Mol Sci 16:13561–13578. doi:10.3390/ijms160613561
Staszak AM, Pawłowski T (2012) Forest tree research in post genomic era. Introduction to systems biology of broadleaves. Dendrobiology 68:113–123
Staszak AM, Pawłowski TA (2014) Proteomic analysis of embryogenesis and the acquisition of seed dormancy in norway maple (Acer platanoides L.). Int J Mol Sci 15:10868–10891. doi:10.3390/ijms150610868
Stone GN, Schonrogge K, Atkinson RJ et al (2002) The population biology of oak gall wasps (Hymenoptera: Cynipidae). Annu Rev Entomol 47:633–668. doi:10.1146/annurev.ento.47.091201.145247
Thomas C, Tholl S, Moes D et al (2009) Actin bundling in plants. Cell Motil Cytoskelet 66:940–957. doi:10.1002/cm.20389
Tian F-X, Zang J-L, Wang T et al (2015) Aldehyde Dehydrogenase gene superfamily in populus: organization and expression divergence between paralogous gene pairs. PLoS One. doi:10.1371/journal.pone.0124669
Tiburcio AF, Altabella T, Bitrián M, Alcázar R (2014) The roles of polyamines during the lifespan of plants: from development to stress. Planta 240:1–18. doi:10.1007/s00425-014-2055-9
Vandenbussche F, Vaseva I, Vissenberg K, Van Der Straeten D (2012) Ethylene in vegetative development: a tale with a riddle. New Phytol 194:895–909. doi:10.1111/j.1469-8137.2012.04100.x
Volkmann D, Baluška F (1999) Actin cytoskeleton in plants: from transport networks to signaling networks. Microsc Res Tech 47:135–154. doi:10.1002/(SICI)1097-0029(19991015)47:2<135:AID-JEMT6>3.0.CO;2-1
Wei Y, Lin M, Oliver DJ, Schnable PS (2009) The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem 10:7. doi:10.1186/1471-2091-10-7
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493. doi:10.1104/pp.126.2.485
Yang Z-D, Lv W-L, Zheng X-L et al (2015) Effect of a gall-inducing psylloid, Pseudophacopteron alstonium Yang et Li (Hemiptera: Phacopteronidae) on defensive chemistry of Alstonia scholaris (L.) R. Br. (Gentianales: Apocynaceae). Turk Entomol Derg (Turk J Entomol) 39:393–400
Zhang S, Shukle R, Mittapalli O et al (2010) The gut transcriptome of a gall midge, Mayetiola destructor. J Insect Physiol 56:1198–1206. doi:10.1016/j.jinsphys.2010.03.021
Acknowledgements
The authors wish to thank the Michał Dadlez group (Mass Spectrometry Laboratory, IBB PAS, Warsaw, Poland) for the MS analyses. The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by the European Regional Development Fund and Innovative Economy, the National Cohesion Strategy of Poland. The work was supported by the Ministry of Science and Higher Education, Poland, Grant Number NN304 210737.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M Hajduch.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pawłowski, T.A., Staszak, A.M., Karolewski, P. et al. Plant development reprogramming by cynipid gall wasp: proteomic analysis. Acta Physiol Plant 39, 114 (2017). https://doi.org/10.1007/s11738-017-2414-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2414-9






