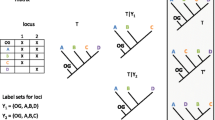
Each informative character, either binary or multistate, corresponds to a tree. A simple example is a binary character absent from taxon A (state 0) and present in taxa BC (state 1). If state 0 is the root, then the corresponding tree is A(BC).
Platnick et al. (1996, p. 250).
Abstract
The possibility of undertaking matrix/optimization-free cladistic analysis is one of the most interesting ideas to emerge in the last few decades from within the field of systematics, particularly in the development of cladistics. The purpose of this paper is to design further opportunities and prospects made possible by eliminating the matrix as the primary source of data representation. The main focus of this paper is to outline a supertree approach that, if combined with the methodology of three-taxon statement analysis (3TA), may be seen as a powerful heuristic alternative to the application of conventional matrix/optimization-based methods used for the analysis of systematic data, and which currently forms the mainstream of contemporary phylogenetics. Using the average consensus technique as an example, we demonstrate explicitly that methods of construction of supertrees may be applied to the array of three-taxon statements (3TS), especially if the latter are represented initially as minimal trees, not as binary matrices, as was originally proposed. The 3TA-average consensus procedure recognizes solely ‘reversal’-based clades and is also free from the potential issues of 3TA, such as the data distortion due to inability to handle putative reversals. Thus the main benefit of this new approach over the traditional one is its accuracy and advantages when implementing the Hennigian views on the cladistic analysis that states that all characters must be a priori polarized before the best fitting tree is found. We also found that the average consensus technique (as well as other median supertree calculation techniques) is purely typological and we stressed that this simple point had never been mentioned before. We proposed that the average consensus of 3TSs (as well as any median consensus of 3TSs) may be viewed as a median type and the extended procedure of the traditional 3TA may be treated as a typology of the relations. The connection between median type and phylogeny may be established only indirectly. The heuristic scientific typology may be derived within a completely metaphysics-free context. Goethe’s idea of “Urphenomenon” and Max Weber’s “Ideal Types” are mentioned as examples of heuristic metaphysical-free typological frameworks.






Similar content being viewed by others
References
Aguirre-Fernández, G., Barnes, L. G., Aranda-Manteca, F. J., & Fernández-Rivera, J. R. (2009). Protoglobicephala mexicana, a new genus and species of Pliocene fossil dolphin (Cetacea; Odontoceti; Delphinidae) from the Gulf of California. Boletín de la Sociedad Geológica Mexicana, 61(2), 245–265.
Ax, P. (1987). The phylogenetic system. The systematization of organisms on the basis of their phylogenesis. Chichester: Wiley.
Baum, B. R. (1992). Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon, 41(1), 3–10.
Bininda-Emonds, O. R. (2014). An introduction to supertree construction (and partitioned phylogenetic analyses) with a view toward the distinction between gene trees and species trees. In L. Z. Garamszegi (Ed.), Modern phylogenetic comparative methods and their application in evolutionary biology (pp. 49–76). Berlin: Springer.
Bininda-Emonds, O. R. P. (2004). The evolution of supertrees. Trends in Ecology & Evolution, 19(6), 315–322.
Brady, R. H. (1982). Theoretical issues and “pattern cladists”. Systematic Zoology, 31(3), 286–291.
Brady, R. H. (1985). On the independence of systematics. Cladistics, 1(2), 113–126.
Brower, A. V. Z. (2015). Transformational and taxic homology revisited. Cladistics, 31(2), 197–201.
Bruen, T. C., & Bryant, D. (2008). Parsimony via consensus. Systematic Biology, 57(2), 251–256.
Bruun, H. H. (2001). Weber on Rickert: From value relation to ideal type. Max Weber Studies, 1(2), 138–160.
Burger, T. (1978). Max Weber’s theory of concept formation: History, laws and ideal types. Duran: Duke University Press.
Cantino, P. D., & de Queiroz, K. (2010). PhyloCode: A phylogenetic code of biological nomenclature. Version 4c.
Cao, N., Zaraguëta-Bagils, R., & Vignes-Lebbe, R. (2007). Hierarchical representation of hypotheses of homology. Geodiversitas, 29(1), 5–15.
Carine, M. A., & Scotland, R. W. (1999). Taxic and transformational homology: Different ways of seeing. Cladistics, 15, 121–129.
Carpenter, J. C. (1987). Cladistics of cladists. Cladistics, 3(4), 363–375.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552.
Chen, D., Eulenstein, O., & Fernandez-Baca, D. (2004). Rainbow: A toolbox for phylogenetic supertree construction and analysis. Bioinformatics, 20(16), 2872–2873.
Cotton, J. A., & Page, R. D. M. (2004). Tangled trees from molecular markers: Reconciling conflict between phylogenies to build molecular supertrees. In O. R. P. Bininda-Emonds (Ed.), Phylogenetic supertrees: Combining information to reveal the tree of life (pp. 107–125). Dordrecht: Springer.
Creevey, C. (2004). Clann: Construction of supertrees and exploration of phylogenomic information from partially overlapping datasets (version 3.0.0), user manual. 3.0 ed. Manchester, Great Britan: The lab of James McInerney. http://chriscreevey.github.io/clann/.
Creevey, C. J., & McInerney, J. O. (2009). Trees from trees: Construction of phylogenetic supertrees using Clann. In D. Posada (Ed.), Bioinformatics for DNA Sequence Analysis, (pp. 139–161). Humana Press-Springer-Nature, Switzerland.
Desper, R., & Gascuel, O. (2002). Fast and accurate phylogeny reconstruction algorithms based on the minimum evolution principle. Journal of Computational Biology, 9(5), 687–705.
Ebach, M. C. (2005). Anschauung and the Archetype: The role of Goethe’s delicate empiricism in comparative biology. Janus Head, 8(1), 254–270.
Ebach, M. C. (2017). “Mehr Licht!” Anschauung and its fading role in Morphology. In: J. F. G. Toni, R. Richter, & P. Schilperoord (Eds.), Evolving morphology (pp. 22–37). Dornach.
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797.
Eldredge, N., & Cracraft, J. (1980). Phylogenetic patterns and the evolutionary process. New York: Columbia University Press.
Farris, J. S. (1997). Cycles. Cladistics, 13(1–2), 131–144.
Farris, J. S., & Kluge, A. G. (1998). A/the brief history of three–taxon analysis. Cladistics, 14(4), 349–362.
Felsenstein, J. (1989). PHYLIP – phylogeny inference package (Version 3.2). Cladistics, 5(2), 164–166.
Felsenstein, J. (2004). Inferring phylogenies (2nd ed.). Sunderland: Sinauer Associates, Inc.
Goloboff, P. A., & Pol, D. (2002). Semi-strict supertrees. Cladistics, 18(5), 514–525.
Goremykin, V. V., Nikiforova, S. V., Biggs, P. J., Zhong, B., Delange, P., Martin, W., et al. (2013). The evolutionary root of flowering plants.. Systematic Biology, 62(1), 50–61.
Gouy, M., Guindon, S., & Gascuel, O. (2010). SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution, 27(2), 221–224.
Grand, A., Corvez, A., Duque Velez, L. M., & Laurin, M. (2013). Phylogenetic inference using discrete characters: Performance of ordered and unordered parsimony and of three-item statements. Biological Journal of the Linnean Society, 110(4), 914–930.
Heincke, F. (1898). Naturgeschichte des Herings. Teil I. Die Lokalformen und die Wanderungen des Heringes in den europaischen Meeren. In Abhandlungen des Deutschen Seefischerei-Vereins (Bd. II). Berlin: Verlag von Otto Sale. https://babel.hathitrust.org/cgi/pt?id=chi.23758345;view=1up;seq=7.
Hennig, W. (1966). Phylogenetic systematics (D. Davis, & R. Zangerl, Trans.). Urbana: University of Illinois Press.
Hobbs, C. R., & Baldwin, B. G. (2013). Asian origin and upslope migration of Hawaiian Artemisia (Compositae-Anthemideae). Journal of Biogeography, 40(3), 442–454.
Kitching, I. J., Forey, P. L., Humphries, C. J., & Williams, D. M. (1998). Cladistics: The theory and practice of parsimony analysis (Vol. 11, 2nd ed.). Oxford: Systematics Association Publication.
Kluge, A. G. (1994). Moving targets and shell games. Cladistics, 10(4), 403–413.
Kluge, A. G., & Farris, J. S. (1999). Taxic homology equals overall similarity. Cladistics, 15(2), 205–212.
Kuo, L.-Y., Qi, X., Ma, H., & Li, F.-W. (2018). Order-level fern plastome phylogenomics: New insights from Hymenophyllales. American Journal of Botany, 105, 1545–1555.
Lapointe, F. J., & Cucumel, G. (1997). The average consensus procedure: Combination of weighted trees containing identical or overlapping sets of taxa. Systematic Biology, 46(2), 306–312.
Lapointe, F. J., & Levasseur, C. (2004). Everything you always wanted to know about the average consensus, and more. In O. R. P. Bininda-Emonds (Ed.), Phylogenetic supertrees: Combining information to reveal the tree of life (pp. 87–105). Dordrecht: Springer.
Lapointe, F. J., Wilkinson, M., & Bryant, D. (2003). Matrix representations with parsimony or with distances: Two sides of the same coin? Systematic Biology, 52(6), 865–868.
Laurin, M., de Queiroz, K., Cantino, P. D., Cellinese, N., & Olmstead, R. (2005). The PhyloCode, types, ranks, and monophyly: A response to Pickett. Cladistics, 21(5), 605–607.
Maddison, W. P., & Maddison, D. R. (2011). Mesquite: A modular system for evolutionary analysis. Version 3.01. Retrieved from http://mesquiteproject.org/.
Mavrodiev, E. V. (2015). Three-taxon analysis can always successfully recognize groups based on putative reversals. PeerJ PrePrints, 3, e1206. https://doi.org/10.7287/peerj.preprints.979v1.
Mavrodiev, E. V. (2016). Dealing with propositions, not with the characters: The ability of three-taxon statement analysis to recognize groups based solely on ‘reversals’, under the maximum-likelihood criteria. Australian Systematic Botany, 29(2), 119–125.
Mavrodiev, E. V., Dell, C., & Schroder, L. (2017). A laid-back trip through the Hennigian forests. PeerJ, 5, e3578, https://doi.org/10.7717/peerj.3578.
Mavrodiev, E. V., & Madorsky, A. (2012). TAXODIUM Version 1.0: A simple way to generate uniform and fractionally weighted three-item matrices from various kinds of biological data. PLoS ONE, 7(11), e48813. https://doi.org/10.1371/journal.pone.0048813.
Mavrodiev, E. V., Martinez-Azorin, M., Dranishnikov, P., & Crespo, M. B. (2014). At least 23 genera instead of one: The case of Iris L. s.l. (Iridaceae). PLoS ONE 9(8), e106459. https://doi.org/10.1371/journal.pone.0106459.
Mavrodiev, E. V., & Yurtseva, O. V. (2017). “A character does not make a genus, but the genus makes the character”: Three-taxon statement analysis and intuitive taxonomy. European Journal of Taxonomy, 377, 1–7.
Mikoleit, G. (2004). Phylogenetische Systematik der Wirbeltiere. Pfeil, Dr. Friedrich.
Nelson, G. (1989). Cladistics and evolutionary models. Cladistics, 5(3), 275–289.
Nelson, G. 1996. Nullius in verba. New York, Self-published.
Nelson, G. (2004). Cladistics: Its arrested development. In D. M. Williams, & P. L. Forey (Eds.), Milestones in systematics (pp. 127–148). Boca Raton: CRC Press.
Nelson, G., & Ladiges, P. Y. (1994). Three-item consensus: Empirical test of fractional weighting. In R. W. Scotland, D. J. Siebert, & D. M. Williams (Eds.), Models in phylogeny reconstruction (Systematics Association, special volume series) (Vol. 52, pp. 193–209). Oxford: Oxford University Press.
Nelson, G., & Ladiges, P. Y. (1992). Information-content and fractional weight of 3-item statements. Systematic Biology, 41(4), 490–494.
Nelson, G., & Platnick, N. (1981). Systematics and biogeography: Cladistics and vicariance. New York: Columbia University Press.
Nelson, G., & Platnick, N. I. (1991). Three-taxon statements—A more precise use of parsimony? Cladistics, 7(4), 351–366.
Nelson, G. J. (1970). Outline of a theory of comparative biology. Systematic Zoology, 19(4), 373–384.
Patterson, C. (1980). Cladistics. Biologist, 27, 234–240.
Platnick, N. I. (1979). Philosophy and the transformation of cladistics. Systematic Zoology, 28(4), 537–546.
Platnick, N. I. (1993). Character optimization and weighting—Differences between the standard and three-taxon approaches to phylogenetic inference. Cladistics, 9(2), 267–272.
Platnick, N. I. (2012). The poverty of the PhyloCode: A reply to de Queiroz and Donoghue. Systematic Biology, 61(2), 360–361.
Platnick, N. I., Humphries, C. J., Nelson, G., & Williams, D. M. (1996). Is Farris optimization perfect?: Three-taxon statements and multiple branching. Cladistics, 12(3), 243–252.
Powers, J. (2013). Finding Ernst Mayr’s Plato. Studies in History and Philosophy of Biological and Biomedical Sciences, 44(4B), 714–723.
Ragan, M. A. (1992). Phylogenetic inference based on matrix representation of trees. Molecular Phylogenetics and Evolution, 1(1), 53–58.
Rambaut, A. (2012). FigTree Version. 1.4.3. Molecular evolution, phylogenetics and epidemiology. Edinburgh, UK. University of Edinburgh, Institute of Evolutionary Biology. Retrieved from http://tree.bio.ed.ac.uk/software/figtree/.
Ranwez, V., Criscuolo, A., & Douzery, E. J. P. (2010). SuperTriplets: A triplet-based supertree approach to phylogenomics. Bioinformatics, 26(12), i115–i123.
Remane, A. (1952). Grundlagen des Natürlichen Systems, der Vergleichenden Anatomie und der Phylogenetik. Theoretische Morphologie und Systematik. Leipzig: Akademische Verlagsgesellschaft Geert & Portig.
Rieppel, O. (2007). The metaphysics of Henning’s phylogenetic systematics: Substance, events and laws of nature. Systematics and Biodiversity, 5(4), 345–360.
Rieppel, O. (2013). Styles of scientific reasoning: Adolf Remane (1898–1976) and the German evolutionary synthesis. Journal of Zoological Systematics and Evolutionary Research, 51, 1–12.
Rieppel, O., Williams, D. M., & Ebach, M. C. (2013). Adolf Naef (1883–1949): On foundational concepts and principles of systematic morphology. Journal of the History of Biology, 46(3), 445–510.
Rineau, V., Grand, A., Zaraguëta-Bagils, R., & Laurin, M. (2015). Experimental systematics: Sensitivity of cladistic methods to polarization and character ordering schemes. Contributions to Zoology, 84(2), 129–148.
Rineau, V., Zaraguëta-Bagils, R., & Laurin, M. (2018). Impact of errors on cladistic inference: Simulation-based comparison between parsimony and three-taxon analysis. Contributions to Zoology, 87(1), 25–40.
Schmitt, M. (2016a). Hennig, Ax, and present-day mainstream cladistics on polarizing characters. Peckiana, 11, 35–42.
Schmitt, M. (2016b). How much of Hennig is in present day cladistics? In D. M. Williams, M. Schmitt, & Q. Wheeler (Eds.), The future of phylogenetic systematics: The legacy of Willi Hennig (Systematics Association, special volume series) (Vol. 86, pp. 115–127). Cambridge: Cambridge University Press.
Siebert, D. J., & Williams, D. M. (1998). Recycled. Cladistics, 14(4), 339–347.
Smirnov, E. (1925). The theory of type and natural system. Zeitschrift fuer Induktive Abstammungs und Vererbungslehre (Berlin), 37, 28–66.
Sokal, R. R. (1962). Typology and empiricism in taxonomy. Journal of Theoretical Biology, 3(2), 230–267.
Sokal, R. R., & Sneath, P. H. A. (1963). Principles of numerical taxonomy. San Francisco: W. H. Freeman.
Stevens, P. F. (1983). Report of third annual Willi Hennig Society meeting. Systematic Zoology, 32(3), 285–291.
Swofford, D. L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0. Sunderland: Sinauer Associates.
Thorley, J. L., & Wilkinson, M. (2003). A view of supertree methods. In Janowitz, M. E. et al. (Eds.), Bioconsensus: DIMACS Working Group Meetings on Bioconsensus: October 25–26, 2000 and October 2–5, 2001, DIMACS Center, vol. 61 (pp. 185–194). American Mathematical Society.
Tremblay, F. (2013). Nicolai Hartmann and the metaphysical foundation of phylogenetic systematics. Biology Theory, 7(1), 56–68.
Waegele, J. W. (2005). Foundations of phylogenetic systematics. München: Pfeil Verlag.
Watkins, J. W. (1952). Ideal types and historical explanation. The British Journal for the Philosophy of Science, 3(9), 22–43.
Weberling, F. (1999). Wilhelm Troll, his work and influence. Systematics and Geography of Plants, 68, 9–24.
Wheeler, W. (1996). Optimization alignment: The end of multiple sequence alignment in phylogenetics? Cladistics, 12(1), 1–9.
Wiesemüller, B., Rothe, H., & Hencke, W. (2003). Phylogenetische Systematik: Eine Einführung. Berlin: Springer.
Wiley, E. O., & Lieberman, B. S. (2011). Phylogenetics: The theory and practice of phylogenetic systematics (2nd ed.). Hoboken: Wiley
Wilkinson, M., Cotton, J. A., Creevey, C., Eulenstein, O., Harris, S. R., Lapointe, F. J., et al. (2005a). The shape of supertrees to come: Tree shape related properties of fourteen supertree methods. Systematic Biology, 54(3), 419–431.
Wilkinson, M., Cotton, J. A., & Thorley, J. L. (2004). The information content of trees and their matrix representations. Systematic Biology, 53, 989–1001.
Wilkinson, M., Pisani, D., Cotton, J. A., & Corfe, I. (2005b). Measuring support and finding unsupported relationships in supertrees. Systematic Biology, 54(5), 823–831.
Williams, D. M. (1994). Combining trees and combining data. Taxon, 43(3), 449–453.
Williams, D. M. (1996). Characters and cladograms. Taxon, 45(2), 275–283.
Williams, D. M. (2002). Precision and parsimony. Taxon, 51(1), 143–149.
Williams, D. M. (2004). Supertrees, components and three-item data. In O. R. P. Bininda-Emonds (Ed.), Phylogenetic supertrees: Combining information to reveal the tree of life (pp. 389–408). Dordrecht: Springer.
Williams, D. M., & Ebach, M. C. (2005). Drowning by numbers: Rereading Nelson’s “Nullius in Verba”. Botanical Review, 71, 415–447.
Williams, D. M., & Ebach, M. C. (2006). The data matrix. Geodiversitas, 28(3), 409–420.
Williams, D. M., & Ebach, M. C. (2008). Foundations of systematics and biogeography. New York: Springer.
Williams, D. M., & Siebert, D. J. (2000). Characters, homology and three-item analysis. In R. W. Scotland & R. T. Pennington (Eds.), Homology and systematics: Coding characters for phylogenetic analysis (Systematics Association, special volume series) (Vol. 58, pp. 183–208). Chapman and Hall: Taylor and Francis.
Winsor, M. P. (2006a). Linnaeus’s biology was not essentialist. Annals of the Missouri Botanical Garden, 93(1), 2–7.
Winsor, M. P. (2006b). The creation of the essentialism story: An exercise in metahistory. History and Philosophy of the Life Sciences, 28(2), 149–174.
Witteveen, J. (2015a). “A temporary oversimplification”: Mayr, Simpson, Dobzhansky, and the origins of the typology/population dichotomy (part 1 of 2). Studies in History and Philosophy of Biological and Biomedical Sciences, 54, 20–33.
Witteveen, J. (2015b). Naming and contingency: The type method of biological taxonomy. Biology & Philosophy, 30(4), 569–586.
Witteveen, J. (2016). “A temporary oversimplification”: Mayr, Simpson, Dobzhansky, and the origins of the typology/population dichotomy (part 2 of 2). Studies in History and Philosophy of Biological and Biomedical Sciences, 57, 96–105.
Yurtseva, O. V., Severova, E. E., & Mavrodiev, E. V. (2017). Persepolium (Polygoneae): A new genus in Polygonaceae based on conventional maximum parsimony and three-taxon statement analyses of a comprehensive morphological dataset. Phytotaxa, 314(2), 151–194.
Zaraguëta-Bagils, R., Ung, V., Grand, A., Vignes-Lebbe, R., Cao, N., & Ducasse, J. (2012). LisBeth: New cladistics for phylogenetics and biogeography. Comptes Rendus Palevol, 11(8), 563–566.
Acknowledgements
The authors wish to thank Prof. Michel Laurin (CNRS/MNHN/UPMC, Sorbonne Universités, Paris, France), Dr. Valentin Rineau (CNRS/MNHN/UPMC, Sorbonne Universités, Paris, France) and an anonymous reviewer for their helpful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Mavrodiev, E.V., Williams, D.M. & Ebach, M.C. On the Typology of Relations. Evol Biol 46, 71–89 (2019). https://doi.org/10.1007/s11692-018-9468-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-018-9468-5