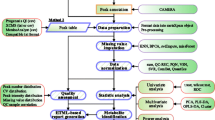
Abstract
LC–MS based global metabolite profiling currently lacks detailed guidelines to demonstrate that the obtained data is of high enough analytical quality. Insufficient data quality may result in the failure to generate a hypothesis, or in the worst case, a false or skewed hypothesis. After assessing the literature, it is apparent that an analytically focused summary and critical discussion related to this subject would be beneficial for both beginners and experts engaged in this field. A particular focus will be placed on data quality, which we here define as the degree to which a set of parameters fulfills predetermined criteria, similar to the established guidelines for targeted analysis. However, several of these parameters are difficult to assess since holistic approaches measure thousands of metabolites in parallel and seldom include predefined knowledge of which metabolites will differ between sample groups. In this review, the following parameters will be discussed in detail: reproducibility, selectivity, certainty of metabolite identification and metabolite coverage. The review systematically describes the generic workflow for LC–MS based global metabolite profiling and highlights how each separate part may affect data quality. The last part of the review describes how data quality can be evaluated as well as identifies areas where additional improvement is needed. In this review, we provide our own analytical opinions in regards to evaluation and, to some extent, improvement of data quality.




Similar content being viewed by others
References
Allwood, J. W., Erban, A., de Koning, S., Dunn, W. B., Luedemann, A., Lommen, A., et al. (2009). Inter-laboratory reproducibility of fast gas chromatography–electron impact–time of flight mass spectrometry (GC–EI–TOF/MS) based plant metabolomics. Metabolomics, 5(4), 479–496. doi:10.1007/s11306-009-0169-z.
Armitage, E. G., Godzien, J., Alonso-Herranz, V., López-Gonzálvez, Á., & Barbas, C. (2015). Missing value imputation strategies for metabolomics data. Electrophoresis, 36, 3050–3060. doi:10.1002/elps.201500352.
Bauer, C., Cramer, R., & Schuchhardt, J. (2011). Data Mining in Proteomics. Methods in Enzymology, 696(1), 93–105. doi:10.1007/978-1-60761-987-1.
Bell, D. S., Cramer, H. M., & Jones, A. D. (2005). Rational method development strategies on a fluorinated liquid chromatography stationary phase: Mobile phase ion concentration and temperature effects on the separation of ephedrine alkaloids. Journal of Chromatography A, 1095(1–2), 113–118. doi:10.1016/j.chroma.2005.08.004.
Benton, H. P., Want, E., Keun, H. C., Amberg, A., Plumb, R. S., Goldfain-Blanc, F., et al. (2012). Intra- and interlaboratory reproducibility of ultra performance liquid chromatography-time-of-flight mass spectrometry for urinary metabolic profiling. Analytical Chemistry, 84(5), 2424–2432. doi:10.1021/ac203200x.
Bijlsma, S., Bobeldijk, I., Verheij, E. R., Ramaker, R., Kochhar, S., Macdonald, I. A., et al. (2006). Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Analytical Chemistry, 78(2), 567–574. doi:10.1021/ac051495j.
Boron, W. F. (2004). Regulation of intracellular pH. Advances in Physiology Education, 28, 160–179. doi:10.1152/advan.00045.2004.
Bowen, B. P., & Northen, T. R. (2010). Dealing with the unknown: Metabolomics and metabolite atlases. Journal of the American Society for Mass Spectrometry, 21(9), 1471–1476. doi:10.1016/j.jasms.2010.04.003.
Brereton, R. G., & Lloyd, G. R. (2014). Partial least squares discriminant analysis: Taking the magic away. Journal of Chemometrics, 28(4), 213–225. doi:10.1002/cem.2609.
Broadhurst, D. I., & Kell, D. B. (2006). Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics, 2(4), 171–196. doi:10.1007/s11306-006-0037-z.
Brodsky, L., Moussaieff, A., Shahaf, N., Aharoni, A., & Rogachev, I. (2010). Evaluation of peak picking quality in LC-MS metabolomics data. Analytical Chemistry, 82(22), 9177–9187. doi:10.1021/ac101216e.
Brown, M., Dunn, W. B., Dobson, P., Patel, Y., Winder, C. L., Francis-McIntyre, S., et al. (2009). Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst, 134(7), 1322–1332. doi:10.1039/b901179j.
Brown, M., Wedge, D. C., Goodacre, R., Kell, D. B., Baker, P. N., Kenny, L. C., et al. (2011). Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics, 27(8), 1108–1112. doi:10.1093/bioinformatics/btr079.
Bruce, S. J., Jonsson, P., Antti, H., Cloarec, O., Trygg, J., Marklund, S. L., & Moritz, T. (2008). Evaluation of a protocol for metabolic profiling studies on human blood plasma by combined ultra-performance liquid chromatography/mass spectrometry: From extraction to data analysis. Analytical Biochemistry, 372(2), 237–249. doi:10.1016/j.ab.2007.09.037.
Bruce, S. J., Tavazzi, I., Rezzi, S., Kochhar, S., & Guy, P. A. (2009). Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography/mass spectrometry. Analytical Chemistry, 81(9), 3285–3296.
Burton, L., Ivosev, G., Tate, S., Impey, G., Wingate, J., & Bonner, R. (2008). Instrumental and experimental effects in LC-MS-based metabolomics. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 871(2), 227–235. doi:10.1016/j.jchromb.2008.04.044.
Bylesjö, M., Rentalainen, M., Cloarec, O., Nicholson, J. K., Holmes, E., & Trygg, J. (2006). OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics, 20(September), 341–351. doi:10.1002/cem.1006.
Calbiani, F., Careri, M., Elviri, L., Mangia, A., & Zagnoni, I. (2006). Matrix effects on accurate mass measurements of low-molecular weight compounds using liquid chromatography-electrospray-quadrupole time-of-flight mass spectrometry. Journal of Mass Spectrometry, 41(3), 289–294. doi:10.1002/jms.984.
Castillo, S., Gopalacharyulu, P., Yetukuri, L., & Orešič, M. (2011). Algorithms and tools for the preprocessing of LC-MS metabolomics data. Chemometrics and Intelligent Laboratory Systems, 108(1), 23–32. doi:10.1016/j.chemolab.2011.03.010.
Coble, J. B., & Fraga, C. G. (2014). Comparative evaluation of preprocessing freeware on chromatography/mass spectrometry data for signature discovery. Journal of Chromatography A, 1358, 155–164. doi:10.1016/j.chroma.2014.06.100.
Coulier, L., Bas, R., Jespersen, S., Verheij, E., van der Werf, M. J., & Hankemeier, T. (2006). Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography–electrospray ionization mass spectrometry. Analytical Chemistry, 78(18), 6573–6582. doi:10.1021/Ac0607616.
Creek, D. J., Dunn, W. B., Fiehn, O., Griffin, J. L., Hall, R. D., Lei, Z., et al. (2014). Metabolite identification: are you sure? And how do your peers gauge your confidence? Metabolomics, 10(3), 350–353. doi:10.1007/s11306-014-0656-8.
Creek, D. J., Jankevics, A., Burgess, K. E. V., Breitling, R., & Barrett, M. P. (2012). IDEOM: An excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics, 28(7), 1048–1049. doi:10.1007/s11306-011-0341-0.
Cuhadar, S., Koseoglu, M., Atay, A., & Dirican, A. (2013). The effect of storage time and freeze-thaw cycles on the stability of serum samples. Biochem Med (Zagreb), 23(1), 70–77.
De Livera, A. M., Dias, D. A., Souza, D. De, Rupasinghe, T., Tull, D. L., Roessner, U., et al. (2012). Normalising and integrating metabolomics data normalising and integrating metabolomics data. Analytical Chemistry, 84, 10768–10776.
Denery, J. R., Nunes, A. A. K., & Dickerson, T. J. (2011). Characterization of differences between blood sample matrices in untargeted metabolomics. Analytical Chemistry, 83, 1040–1047.
Dettmer, K., Aronov, P. A., & Hammock, B. D. (2012). Mass spectrometry-based metabolomics. Mass Spectrometry Reviews, 29(6), 997–1003. doi:10.1016/j.biotechadv.2011.08.021.Secreted.
Di Guida, R., Engel, J., Allwood, J. W., Weber, R. J. M., Jones, M. R., Sommer, U., et al. (2016). Non-targeted UHPLC-MS metabolomic data processing methods: A comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics, 12(5), 93. doi:10.1007/s11306-016-1030-9.
Dieterle, F., Ross, A., Schlotterbeck, G., & Senn, H. (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Analytical Chemistry, 78(13), 4281–4290. doi:10.1021/ac051632c.
Draisma, H. H. M., Reijmers, T. H., & Van Der Kloet, F. (2010). Equating, or correction for between-block effects with application to body fluid LC–MS and NMR metabolomics datasets. Analytical Chemistry, 82(3), 1039–1046.
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols, 6(7), 1060–1083. doi:10.1038/nprot.2011.335.
Dunn, W. B., Erban, A., Weber, R. J. M., Creek, D. J., Brown, M., Breitling, R., et al. (2013). Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics, 9(suppl. 1), 44–66. doi:10.1007/s11306-012-0434-4.
Dunn, W. B., Wilson, I. D., Nicholls, A. W., & Broadhurst, D. (2012). The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis, 4(18), 2249–2264. doi:10.4155/bio.12.204.
Eliasson, M., Ränner, S., Madsen, R., Donten, M. A., Marsden-Edwards, E., Moritz, T., et al. (2012). Strategy for optimizing LC–MS data processing in metabolomics: A design of experiments approach. Analytical Chemistry, 84(15), 6869–6876. doi:10.1016/j.ijpharm.2011.11.009.
EMA. Guideline on bioanalytical method validation., EMA Guideline (2012). EMEA/CHMP/EWP/192217/2009.
Engskog, M., Björklund, M., Haglöf, J., Arvidsson, T., Shoshan, M., & Pettersson, C. (2015). Metabolic profiling of epithelial ovarian cancer cell lines: Evaluation of harvesting protocols for profiling using NMR spectroscopy. Bioanalysis, 7(2), 157–166.
Eriksson, L., Byrne, T., Johansson, E., Trygg, J., & Vikström, C. (2013). Centering and Scaling. In Multi- and Megavariate Data Analysis (3rd ed., pp. 243–254). Malmö: MKS Umetrics AB.
Fiehn, O. (2002). Metabolomics—The link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171. doi:10.1023/A:1013713905833.
Food and Drug Administration. (2001). Guidance for industry: Bioanalytical method validation. U.S. Department of Health and Human Services. doi:http://www.labcompliance.de/documents/FDA/FDA-Others/Laboratory/f-507-bioanalytical-4252fnl.pdf.
Food and Drug Administration. (2013). Guidance for industry bioanalytical method validation guidance for industry bioanalytical method validation. U.S. Department of Health and Human Services. doi:http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm368107.pdf.
Fura, A., Harper, T. W., Zhang, H., Fung, L., & Shyu, W. C. (2003). Shift in pH of biological fluids during storage and processing: Effect on bioanalysis. Journal of Pharmaceutical and Biomedical Analysis, 32(3), 513–522. doi:10.1016/S0731-7085(03)00159-6.
Gertsman, I., Gangoiti, J., & Barshop, B. (2014). Validation of a dual LC-HRMS platform for clinical metabolic diagnosis in serum, bridging quantitative analysis and untargeted metabolomics. Metabolomics, 10(2), 312–323. doi:10.1016/j.biotechadv.2011.08.021.Secreted.
Gika, H. G., Macpherson, E., Theodoridis, G. A., & Wilson, I. D. (2008). Evaluation of the repeatability of ultra-performance liquid chromatography-TOF-MS for global metabolic profiling of human urine samples. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 871(2), 299–305. doi:10.1016/j.jchromb.2008.05.048.
Gika, H. G., Theodoridis, G. A., Earll, M., & Wilson, I. D. (2012a). A QC approach to the determination of day-to-day reproducibility and robustness of LC–MS methods for global metabolite profiling in metabonomics/metabolomics. Bioanalysis, 4(18), 2239–2247. doi:10.4155/bio.12.212.
Gika, H., Theodoridis, G., Mattivi, F., Vrhovsek, U., & Pappa-Louisi, A. (2012b). Retention prediction of a set of amino acids under gradient elution conditions in hydrophilic interaction liquid chromatography. Journal of Separation Science, 35(3), 376–383. doi:10.1002/jssc.201100795.
Gika, H. G., Theodoridis, G. A., Plumb, R. S., & Wilson, I. D. (2014a). Current practice of liquid chromatography–mass spectrometry in metabolomics and metabonomics. Journal of Pharmaceutical and Biomedical Analysis, 87, 12–25. doi:10.1016/j.jpba.2013.06.032.
Gika, H. G., Theodoridis, G. A., Wingate, J. E., & Wilson, I. D. (2007). Within-day reproducibility of an HPLC–MS-based method for metabonomic analysis: Application to human urine research articles. Journal of Proteome Research, 6(8), 3291–3303.
Gika, H. G., Wilson, I. D., & Theodoridis, G. A. (2014b). LC-MS-based holistic metabolic profiling. Problems, limitations, advantages, and future perspectives. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 966, 1–6. doi:10.1016/j.jchromb.2014.01.054.
Gika, H. G., Zisi, C., Theodoridis, G., & Wilson, I. D. (2016). Protocol for quality control in metabolic profiling of biological fluids by U(H)PLC-MS. Journal of Chromatography B, 1008, 15–25. doi:10.1016/j.jchromb.2015.10.045.
Goeddel, L., & Patti, G. (2012). Maximizing the value of metabolomic data. Bioanalysis, 4(18), 2199–2201. doi:10.4155/bio.12.210.
Goodacre, R. (2007). Metabolomics of a superorganism. The Journal of Nutrition, 137(suppl. 1), 259S–266S.
Gromski, P. S., Muhamadali, H., Ellis, D. I., Xu, Y., Correa, E., Turner, M. L., & Goodacre, R. (2015). A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Analytica Chimica Acta, 879, 10–23. doi:10.1016/j.aca.2015.02.012.
Gromski, P. S., Xu, Y., Kotze, H. L., Correa, E., Ellis, D. I., Armitage, E. G., et al. (2014). Influence of missing values substitutes on multivariate analysis of metabolomics data. Metabolites, 4(2), 433–452. doi:10.3390/metabo4020433.
Gürdeniz, G., Kristensen, M., Skov, T., & Dragsted, L. O. (2012). The effect of LC–MS data preprocessing methods on the selection of plasma biomarkers in fed versus fasted rats. Metabolites, 2(1), 77–99. doi:10.3390/metabo2010077.
Hebels, D. G. A., Georgiadis, P., Keun, H. C., Athersuch, T. J., Vineis, P., Vermeulen, R., et al. (2013). Performance in omics analyses of blood samples in long-term storage: Opportunities for the exploitation of existing biobanks in environmental. Environmental Health Perspectives, 480(4), 480–487.
Hendriks, G., Uges, D. R., & Franke, J. P. (2007). Reconsideration of sample pH adjustment in bioanalytical liquid-liquid extraction of ionisable compounds. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 853(1–2), 234–241. doi:10.1016/j.jchromb.2007.03.017.
Hendriks, M. M. W. B., van Eeuwijk, F. A., Jellema, R. H., Westerhuis, J. A., Reijmers, T. H., Hoefsloot, H. C. J., & Smilde, A. K. (2011). Data-processing strategies for metabolomics studies. TrAC—Trends in Analytical Chemistry, 30(10), 1685–1698. doi:10.1016/j.trac.2011.04.019.
Hrydziuszko, O., & Viant, M. R. (2012). Missing values in mass spectrometry based metabolomics: An undervalued step in the data processing pipeline. Metabolomics, 8, 161–174. doi:10.1007/s11306-011-0366-4.
Ismaiel, O., Zhang, T., Jenkins, R., & Karnes, H. T. (2011). Determination of octreotide and assessment of matrix effects in human plasma using ultra high performance liquid chromatography-tandem mass spectrometry. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 879(22), 2081–2088. doi:10.1016/j.jchromb.2011.05.039.
Issaq, H. J., Waybright, T. J., & Veenstra, T. D. (2011). Cancer biomarker discovery: Opportunities and pitfalls in analytical methods. Electrophoresis, 32(9), 967–975. doi:10.1002/elps.201000588.
Ivanisevic, J., Zhu, Z. J., Plate, L., Tautenhahn, R., Chen, S., O’Brien, P. J., et al. (2013). Toward’Omic scale metabolite profiling: A dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Analytical Chemistry, 85(14), 6876–6884. doi:10.1021/ac401140h.
Jackson, J. E. (1991). A user’s guide to principal components. New York: Wiley. doi:10.1002/0471725331.
Jørgenrud, B., Jäntti, S. S., Mattila, I., Pöhö, P. P., Rønningen, K. S., Yki-Järvinen, H., et al. (2015). The influence of sample collection methodology and sample preprocessing on the blood metabolic profile. Bioanalysis, 7(8), 991–1006. doi:10.4155/bio.15.16.
Kamlage, B., Maldonado, S. G., Bethan, B., Peter, E., Schmitz, O., Liebenberg, V., & Schatz, P. (2014). Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clinical Chemistry, 60(2), 399–412. doi:10.1373/clinchem.2013.211979.
Kamleh, M. A., Ebbels, T. M. D., Spagou, K., Masson, P., & Want, E. J. (2012). Optimizing the use of quality control samples for signal drift correction in large-scale urine metabolic profiling studies. Analytical Chemistry, 84, 2670–2677.
Katajamaa, M., Miettinen, J., & Orešič, M. (2006). MZmine: Toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics, 22(5), 634–636. doi:10.1093/bioinformatics/btk039.
Kell, D. B. (2004). Metabolomics and systems biology: Making sense of the soup. Current Opinion in Microbiology, 7(3), 296–307. doi:10.1016/j.mib.2004.04.012.
Keun, H. C., Ebbels, T. M. D., Antti, H., Bollard, M. E., Beckonert, O., Holmes, E., et al. (2003). Improved analysis of multivariate data by variable stability scaling: Application to NMR-based metabolic profiling. Analytica Chimica Acta, 490(1–2), 265–276. doi:10.1016/S0003-2670(03)00094-1.
Kind, T., & Fiehn, O. (2010). Advances in structure elucidation of small molecules using mass spectrometry. Bioanalytical Reviews, 2(1), 23–60. doi:10.1007/s12566-010-0015-9.
Kirwan, J. A., Broadhurst, D. I., Davidson, R. L., & Viant, M. R. (2013). Characterising and correcting batch variation in an automated direct infusion mass spectrometry (DIMS) metabolomics workflow. Analytical and Bioanalytical Chemistry, 405(15), 5147–5157. doi:10.1007/s00216-013-6856-7.
Kloos, D. P., Lingeman, H., Niessen, W. M. A., Deelder, A. M., Giera, M., & Mayboroda, O. A. (2013). Evaluation of different column chemistries for fast urinary metabolic profiling. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 927, 90–96. doi:10.1016/j.jchromb.2013.02.017.
Kuhl, C., Tautenhahn, R., Böttcher, C., Larson, T., & Neumann, S. (2012). CAMERA: An integrated strategy for compound spectra extraction and annotation of LC/MS data sets. Analytical Chemistry, 84(1), 283–289. doi:10.1021/ac202450g.
Kuligowski, J., Sanchez-Illana, A., Sanjuan-Herraez, D., Vento, M., & Quintas, G. (2015). Intra-batch effect correction in liquid chromatography-mass spectrometry using quality control samples and support vector regression (QC-SVRC). Analyst, 140(22), 7810–7817. doi:10.1039/c5an01638j.
Kultima, K., Nilsson, A., Scholz, B., Rossbach, U. L., Fälth, M., & Andrén, P. E. (2009). Development and evaluation of normalization methods for label-free relative quantification of endogenous peptides. Molecular & Cellular Proteomics: MCP, 8(10), 2285–2295. doi:10.1074/mcp.M800514-MCP200.
Lahaie, M., Mess, J.-N., Furtado, M., & Garofolo, F. (2010). Elimination of LC–MS/MS matrix effect due to phospholipids using specific solid-phase extraction elution conditions. Bioanalysis, 2(6), 1011–1021. doi:10.4155/bio.10.65.
León, Z., García-Cañaveras, J. C., Donato, M. T., & Lahoz, A. (2013). Mammalian cell metabolomics: Experimental design and sample preparation. Electrophoresis, 34(19), 2762–2775. doi:10.1002/elps.201200605.
Lorenz, M. A., Burant, C. F., & Kennedy, R. T. (2011). Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical Chemistry, 83(9), 3406–3414. doi:10.1021/ac103313x.
Lu, W., Clasquin, M. F., Melamud, E., Amador-Noguez, D., Caudy, A. A., & Rabinowitz, J. D. (2011). NIH public access. Analytical Chemistry, 82(8), 3212–3221. doi:10.1021/ac902837x.Metabolomic.
Madsen, R., Lundstedt, T., & Trygg, J. (2010). Chemometrics in metabolomics—A review in human disease diagnosis. Analytica Chimica Acta, 659(1–2), 23–33. doi:10.1016/j.aca.2009.11.042.
Martano, G., Delmotte, N., Kiefer, P., Christen, P., Kentner, D., Bumann, D., & Vorholt, J. A. (2014). Fast sampling method for mammalian cell metabolic analyses using liquid chromatography–mass spectrometry. Nature Protocols, 10(1), 1–11. doi:10.1038/nprot.2014.198.
Martin, J.-C., Maillot, M., Mazerolles, G., Verdu, A., Lyan, B., Migné, C., et al. (2015). Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics, 11(4), 807–821. doi:10.1007/s11306-014-0740-0.
Michopoulos, F., Lai, L., Gika, H., Theodoridis, G., & Wilson, I. (2009). UPLC MS based analysis of human plasma for metabonomics using solvent precipitation or solid phase extraction. Journal of Proteome Research, 8(4), 2114–2121. doi:10.1021/pr801045q.
Moco, S., Vervoort, J., Moco, S., Bino, R. J., De Vos, R. C. H., & Bino, R. (2007). Metabolomics technologies and metabolite identification. TrAC —Trends in Analytical Chemistry, 26(9), 855–866. doi:10.1016/j.trac.2007.08.003.
Naz, S., García, A., & Barbas, C. (2013a). Multiplatform analytical methodology for metabolic fingerprinting of lung tissue. Analytical Chemistry, 85(22), 10941–10948. doi:10.1021/ac402411n.
Naz, S., Garcia, A., Rusak, M., & Barbas, C. (2013b). Method development and validation for rat serum fingerprinting with CE-MS: Application to ventilator-induced-lung-injury study. Analytical and Bioanalytical Chemistry, 405(14), 4849–4858. doi:10.1007/s00216-013-6882-5.
Naz, S., Vallejo, M., García, A., & Barbas, C. (2014). Method validation strategies involved in non-targeted metabolomics. Journal of Chromatography A, 1353, 99–105. doi:10.1016/j.chroma.2014.04.071.
Nicholson, J. K., & Lindon, J. C. (2008). Metabonomics. Nature, 455(October), 1054–1056.
Nicholson, J. K., Lindon, J. C., & Holmes, E. (1999). “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica; The Fate of Foreign Compounds in Biological Systems, 29(11), 1181–1189. doi:10.1080/004982599238047.
Nilsson, L. B. (2013). The bioanalytical challenge of determining unbound concentration and protein binding for drugs. Bioanalysis, 5(24), 3033–3050. doi:10.4155/bio.13.274.
Nilsson, L. B., & Schmidt, S. (2001). Simultaneous determination of total and free drug plasma concentrations combined with batch-wise pH-adjustment for the free concentration determinations. Journal of Pharmaceutical and Biomedical Analysis, 24(5–6), 921–927. doi:10.1016/S0731-7085(00)00560-4.
Ogata, H., Goto, S., Sato, K., Fujubuchi, W., Bono, H., & Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 27(1), 29–34.
Paglia, G., Magnúsdóttir, M., Thorlacius, S., Sigurjónsson, Ó. E., Gudmundsson, S., Palsson, B., & Thiele, I. (2012). Intracellular metabolite profiling of platelets: Evaluation of extraction processes and chromatographic strategies. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 898, 111–120. doi:10.1016/j.jchromb.2012.04.026.
Pandher, R., Ducruix, C., Eccles, S. A., & Raynaud, F. I. (2009). Cross-platform Q-TOF validation of global exo-metabolomic analysis: Application to human glioblastoma cells treated with the standard PI 3-Kinase inhibitor LY294002. Journal of Chromatography, B: Analytical Technologies in the Biomedical and Life Sciences, 877(13), 1352–1358. doi:10.1016/j.jchromb.2008.12.001.
Pedreschi, R., Hertog, M. L. A. T. M., Carpentier, S. C., Lammertyn, J., Robben, J., Noben, J. P., et al. (2008). Treatment of missing values for multivariate statistical analysis of gel-based proteomics data. Proteomics, 8(7), 1371–1383. doi:10.1002/pmic.200700975.
Pereira, H., Martin, J.-F., Joly, C., Sébédio, J. L., & Pujos-Guillot, E. (2010). Development and validation of a UPLC/MS method for a nutritional metabolomic study of human plasma. Metabolomics, 6(2), 207–218. doi:10.1007/s11306-009-0188-9.
Phinney, K. W., Ballihaut, G., Bedner, M., Benford, B. S., Camara, J. E., Christopher, S. J., et al. (2013). Development of a standard reference material for metabolomics research. Analytical Chemistry, 85(24), 11732–11738. doi:10.1021/ac402689t.
Pinto, J., Domingues, M. R. M., Galhano, E., Pita, C., Almeida, M. D. C., Carreira, I. M., & Gil, A. M. (2014). Human plasma stability during handling and storage: Impact on NMR metabolomics. The Analyst, 139(5), 1168–1177. doi:10.1039/c3an02188b.
Psychogios, N., Hau, D. D., Peng, J., Guo, A. C., Mandal, R., Bouatra, S., et al. (2011). The human serum metabolome. PLoS One, 6(2), e16957. doi:10.1371/journal.pone.0016957.
Qi, X., Zhang, Y., Gao, J., Chen, T., Zhao, A., Yan, Y., & Jia, W. (2011). Metabolite profiling of hemodialysate using gas chromatography time-of-flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis, 55(5), 1142–1147. doi:10.1016/j.jpba.2011.04.001.
Rafiei, A., & Sleno, L. (2014). Comparison of peak-picking workflows for untargeted liquid chromatography/high-resolution mass spectrometry metabolomics data analysis. Rapid Communications in Mass Spectrometry, 29(1), 119–127. doi:10.1002/rcm.7094.
Ramakrishnan, P., Nair, S., & Rangiah, K. (2016). A method for comparative metabolomics in urine using high resolution mass spectrometry. Journal of Chromatography A, 1443, 83–92. doi:10.1016/j.chroma.2016.02.080.
Ramautar, R., & de Jong, G. J. (2014). Recent developments in liquid-phase separation techniques for metabolomics. Bioanalysis, 6, 1011–1026. doi:10.4155/bio.14.51.
Rico, E., González, O., Blanco, M. E., & Alonso, R. M. (2014). Evaluation of human plasma sample preparation protocols for untargeted metabolic profiles analyzed by UHPLC-ESI-TOF-MS. Analytical and Bioanalytical Chemistry, 406(29), 7641–7652. doi:10.1007/s00216-014-8212-y.
Robert, O., Sabatier, J., Desoubzdanne, D., Lalande, J., Balayssac, S., Gilard, V., et al. (2011). pH optimization for a reliable quantification of brain tumor cell and tissue extracts with (1)H NMR: focus on choline-containing compounds and taurine. Analytical and Bioanalytical Chemistry, 399(2), 987–999. doi:10.1007/s00216-010-4321-4.
Rusilowicz, M., Dickinson, M., Charlton, A., O’Keefe, S., & Wilson, J. (2016). A batch correction method for liquid chromatography–mass spectrometry data that does not depend on quality control samples. Metabolomics, 12(3), 1–11. doi:10.1007/s11306-016-0972-2.
Saccenti, E., Hoefsloot, H. C. J., Smilde, A. K., Westerhuis, J. A., & Hendriks, M. M. W. B. (2014). Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics, 10(3), 361–374. doi:10.1007/s11306-013-0598-6.
Salek, R. M., Steinbeck, C., Viant, M. R., Goodacre, R., & Dunn, W. B. (2013). The role of reporting standards for metabolite annotation and identification in metabolomic studies. GigaScience, 2(1), 13. doi:10.1186/2047-217X-2-13.
Sana, T. R., Roark, J. C., Li, X., Waddell, K., & Fischer, S. M. (2008). Molecular formula and METLIN personal metabolite database matching applied to the identification of compounds generated by LC/TOF-MS. Journal of Biomolecular Techniques, 19(4), 258–266.
Sangster, T., Major, H., Plumb, R., Wilson, A. J., & Wilson, I. D. (2006). A pragmatic and readily implemented quality control strategy for HPLC-MS and GC–MS-based metabonomic analysis. The Analyst, 131(10), 1075–1078. doi:10.1039/b604498k.
Sarafian, M. H., Gaudin, M., Lewis, M. R., Martin, F. P., Holmes, E., Nicholson, J. K., & Dumas, M. E. (2014). Objective set of criteria for optimization of sample preparation procedures for ultra-high throughput untargeted blood plasma lipid profiling by ultra performance liquid chromatography–mass spectrometry. Analytical Chemistry, 86(12), 5766–5774. doi:10.1021/ac500317c.
Scheel, I., Aldrin, M., Glad, I. K., Sørum, R., Lyng, H., & Frigessi, A. (2005). The influence of missing value imputation on detection of differentially expressed genes from microarray data. Bioinformatics, 21(23), 4272–4279. doi:10.1093/bioinformatics/bti708.
Scheltema, R. A., Jankevics, A., Jansen, R. C., Swertz, M. A., & Breitling, R. (2011). PeakML/mzMatch: A file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Analytical Chemistry, 83(7), 2786–2793. doi:10.1021/ac2000994.
Simón-Manso, Y., Lowenthal, M. S., Kilpatrick, L. E., Sampson, M. L., Telu, K. H., Rudnick, P. A., et al. (2013). Metabolite profiling of a NIST standard reference material for human plasma (SRM 1950): GC–MS, LC–MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Analytical Chemistry, 85(24), 11725–11731. doi:10.1021/ac402503m.
Smilde, A. K., Van der Werf, M. J., & Bijlsma, S. (2005). Fusion of mass spectrometry-based metabolomics data. Analytical Chemistry, 77(20), 6729. papers3://publication/uuid/D4413DC1-F642-419B-9706-6E027D8014A8.
Smilde, A. K., van der Werf, M. J., Schaller, J.-P., & Kistemaker, C. (2009). Characterizing the precision of mass-spectrometry-based metabolic profiling platforms. The Analyst, 134(11), 2281. doi:10.1039/b902242b.
Smith, C., Elizabeth, J., O’Maille, G., Abagyan, R., & Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. ACS Publications, 78(3), 779–787.
Spagou, K., Tsoukali, H., Raikos, N., Gika, H., Wilson, I. D., & Theodoridis, G. (2010). Hydrophilic interaction chromatography coupled to MS for metabonomic/metabolomic studies. Journal of Separation Science, 33(6–7), 716–727. doi:10.1002/jssc.200900803.
Sud, M., Fahy, E., Cotter, D., Brown, A., Dennis, E. A., Glass, C. K., et al. (2007). LMSD: LIPID MAPS structure database. Nucleic Acids Research, 35(suppl. 1), 527–532. doi:10.1093/nar/gkl838.
Sumner, L. W., Samuel, T., Noble, R., Gmbh, S. D., Barrett, D., Beale, M. H., & Hardy, N. (2007). Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics, 3(3), 211–221. doi:10.1007/s11306-007-0082-2.Proposed.
Sysi-Aho, M., Katajamaa, M., Yetukuri, L., & Oresic, M. (2007). Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics, 8, 93. doi:10.1186/1471-2105-8-93.
Szymańska, E., Saccenti, E., Smilde, A. K., & Westerhuis, J. A. (2012). Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics, 8(1), 3–16. doi:10.1007/s11306-011-0330-3.
T’Kindt, R., Alaerts, G., Heyden, Y. Vander, Deforce, D., & Van Bocxlaer, J. (2007). Broad-spectrum separations in metabolomics using enhanced polar LC stationary phases: A dedicated evaluation using plant metabolites. Journal of Separation Science, 30(13), 2002–2011. doi:10.1002/jssc.200700077.
Taguchi, R., Nishijima, M., & Shimizu, T. (2007). Basic analytical systems for lipidomics by mass spectrometry in Japan. Methods in Enzymology, 432(07), 185–211. doi:10.1016/S0076-6879(07)32008-9.
Teahan, O., Gamble, S., Holmes, E., Waxman, J., Nicholson, J. K., Bevan, C., & Keun, H. C. (2006). Impact of analytical bias in metabonomic studies of human blood serum and plasma. Analytical Chemistry, 78(13), 4307–4318. doi:10.1021/ac051972y.
Telu, K. H., Yan, X., Wallace, W. E., Stein, S. E., & Simón-Manso, Y. (2016). Analysis of human plasma metabolites across different liquid chromatography/mass spectrometry platforms: Cross-platform transferable chemical signatures. Rapid Communications in Mass Spectrometry, 30(5), 581–593. doi:10.1002/rcm.7475.
Teng, Q., Huang, W., Collette, T. W., Ekman, D. R., & Tan, C. (2009). A direct cell quenching method for cell-culture based metabolomics. Metabolomics, 5(2), 199–208. doi:10.1007/s11306-008-0137-z.
Theodoridis, G. A., Gika, H. G., Want, E. J., & Wilson, I. D. (2012). Liquid chromatography-mass spectrometry based global metabolite profiling: A review. Analytica Chimica Acta, 711, 7–16. doi:10.1016/j.aca.2011.09.042.
Trygg, J., Holmes, E., & Lundstedt, T. (2007). Chemometrics in metabonomics. Journal of Proteome Research, 6(2), 469–479. doi:10.1021/pr060594q.
Tulipani, S., Llorach, R., Urpi-Sarda, M., & Andres-Lacueva, C. (2013). Comparative analysis of sample preparation methods to handle the complexity of the blood fluid metabolome: When less is more. Analytical Chemistry, 85(1), 341–348. doi:10.1021/ac302919t.
Tulipani, S., Mora-Cubillos, X., Jáuregui, O., Llorach, R., García-Fuentes, E., Tinahones, F. J., & Andres-Lacueva, C. (2015). New and vintage solutions to enhance the plasma metabolome coverage by LC-ESI-MS untargeted metabolomics. The not-so-simple process of method performance evaluation. Analytical Chemistry, 87(5), 2639–2647. doi:10.1021/ac503031d.
van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K., & van der Werf, M. J. (2006). Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics, 7, 142. doi:10.1186/1471-2164-7-142.
Van Der Kloet, F. M., Bobeldijk, I., Verheij, E. R., & Jellema, R. H. (2009). Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. Journal of Proteome Research, 8(11), 5132–5141. doi:10.1021/pr900499r.
Veselkov, K. A., Vingara, L. K., Masson, P., Robinette, S. L., Want, E., Li, J. V., et al. (2011). Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Analytical Chemistry, 83, 5864–5872.
Viant, M. R., Bearden, D. W., Bundy, J. G., Burton, I. W., Collette, T. W., Ekman, D. R., et al. (2008). International NMR-based environmental metabolomics intercomparison exercise. Environmental Science and Technology, 43(1), 219–225. doi:10.1021/es802198z.
Vorkas, P. A., Isaac, G., Anwar, M. A., Davies, A. H., Want, E. J., & Holmes, E. (2015). Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: Application to cardiovascular disease. Analytical Chemistry, 87(8), 4184–4193. doi:10.1021/ac503775m.
Vuckovic, D. (2012). Current trends and challenges in sample preparation for global metabolomics using liquid chromatography-mass spectrometry. Analytical and Bioanalytical Chemistry, 403(6), 1523–1548. doi:10.1007/s00216-012-6039-y.
Want, E. J. (2009). Challenges in applying chemometrics to LC-MS-based global metabolite profile data. Bioanalysis, 1(4), 805–819. doi:10.4155/bio.09.64.
Want, E. J., & Masson, P. (2011). Processing and analysis of GC/LC-MS-based metabolomics data. Methods in Molecular Biology, 708(4), 321–334. doi:10.1007/978-1-61737-985-7.
Want, E. J., Wilson, I. D., Gika, H., Theodoridis, G., Plumb, R. S., Shockcor, J., et al. (2010). Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocols, 5(6), 1005–1018. doi:10.1038/nprot.2010.50.
Ward, J. L., Baker, J. M., Miller, S. J., Deborde, C., Maucourt, M., Biais, B., et al. (2010). An inter-laboratory comparison demonstrates that [1H]-NMR metabolite fingerprinting is a robust technique for collaborative plant metabolomic data collection. Metabolomics, 6(2), 263–273. doi:10.1007/s11306-010-0200-4.
Wedge, D. C., Allwood, J. W., Dunn, W. B., Vaughan, A. A., Simpson, K., Brown, M., et al. (2011). Is serum or plasma more appropriate for inter-subject assessment in patients with small-cell lung cancer. Analytical Chemistry, 83, 6689–6697.
Wehrens, R., Jos Hageman, B. A., Fred van Eeuwijk, B., Rik Kooke, B., Pádraic Flood, B. J., Erik Wijnker, B., et al. (2016). Improved batch correction in untargeted MS-based metabolomics. Metabolomics,. doi:10.1007/s11306-016-1015-8.
Westerhuis, J. A., Hoefsloot, H. C. J., Smit, S., Vis, D. J., Smilde, A. K., Velzen, E. J. J., et al. (2008a). Assessment of PLSDA cross validation. Metabolomics, 4(1), 81–89. doi:10.1007/s11306-007-0099-6.
Westerhuis, J. A., van Velzen, E. J. J., Hoefsloot, H. C. J., & Smilde, A. K. (2008b). Discriminant Q2 (DQ2) for improved discrimination in PLSDA models. Metabolomics, 4(4), 293–296. doi:10.1007/s11306-008-0126-2.
Westerhuis, J. A., van Velzen, E. J. J., Hoefsloot, H. C. J., & Smilde, A. K. (2010). Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metabolomics, 6(1), 119–128. doi:10.1007/s11306-009-0185-z.
Wheelock, Å. M., & Wheelock, C. E. (2013). Trials and tribulations of’omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Molecular BioSystems, 9(11), 2589–2596. doi:10.1039/c3mb70194h.
Whiley, L., Godzien, J., Ruperez, F. J., Legido-Quigley, C., & Barbas, C. (2012). In-vial dual extraction for direct LC-MS analysis of plasma for comprehensive and highly reproducible metabolic fingerprinting. Analytical Chemistry, 84(14), 5992–5999. doi:10.1021/ac300716u.
Wishart, D. S. (2009). Computational strategies for metabolite identification in metabolomics. Bioanalysis, 1(9), 1579–1596. doi:10.4155/bio.09.138.
Wishart, D. S. (2011). Advance in metabolite identification. Bioanalysis, 3(15), 1769–1782.
Wishart, D. S., Jewison, T., Guo, A. C., Wilson, M., Knox, C., Liu, Y., et al. (2013). HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Research, 41(D1), 801–807. doi:10.1093/nar/gks1065.
Wishart, D. S., Knox, C., Guo, A. C., Eisner, R., Young, N., Gautam, B., et al. (2009). HMDB: A knowledgebase for the human metabolome. Nucleic Acids Research, 37(suppl. 1), 603–610. doi:10.1093/nar/gkn810.
Wold, S., Sjöström, M., & Eriksson, L. (2001). PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems, 58(2), 109–130. doi:10.1016/S0169-7439(01)00155-1.
Xia, J., Psychogios, N., Young, N., & Wishart, D. S. (2009). MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Research, 37(suppl. 2), 652–660. doi:10.1093/nar/gkp356.
Xia, J., & Wishart, D. S. (2011). Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nature Protocols, 6(6), 743–760. doi:10.1038/nprot.2011.319.
Yang, W., Chen, Y., Xi, C., Zhang, R., Song, Y., Zhan, Q., et al. (2013). Liquid chromatography−tandem mass spectrometry-based plasma metabonomics delineate the effect of metabolites’ stability on reliability of potential biomarkers. Analytical Chemistry, 85, 2606–2610.
Yang, J., Zhao, X., Lu, X., Lin, X., & Xu, G. (2015). A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis. Frontiers in Molecular Biosciences, 2(February), 4. doi:10.3389/fmolb.2015.00004.
Yin, P., Lehmann, R., & Xu, G. (2015). Effects of pre-analytical processes on blood samples used in metabolomics studies. Analytical and Bioanalytical Chemistry, 407, 4879–4892. doi:10.1007/s00216-015-8565-x.
Yu, Z., Kastenmüller, G., He, Y., Belcredi, P., Möller, G., Prehn, C., et al. (2011). Differences between human plasma and serum metabolite profiles. PLoS One, 6(7), 1–6. doi:10.1371/journal.pone.0021230.
Zelena, E., Dunn, W. B., Broadhurst, D., Francis-McIntyre, S., Carroll, K. M., Begley, P., et al. (2009). Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Analytical Chemistry, 81(4), 1357–1364. doi:10.1021/ac8019366.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Mikael K. R. Engskog, Jakob Haglöf, Torbjörn Arvidsson and Curt Pettersson declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Engskog, M.K.R., Haglöf, J., Arvidsson, T. et al. LC–MS based global metabolite profiling: the necessity of high data quality. Metabolomics 12, 114 (2016). https://doi.org/10.1007/s11306-016-1058-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-016-1058-x