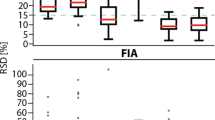
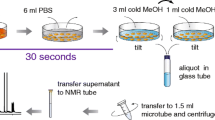
Abstract
Analysis of the metabolome can be sample and cell dependent. In this study, we compared two conventional pre-treatment approaches (trypsinization and cell scraping) in three adherently grown mammalian cell lines (two breast cancer cell lines MDA-MB-436, MCF-7 and an endothelial cell line—HMEC-1). We report experimental evidence, for the first time, demonstrating that metabolite leakage occurs with both treatments, and that the cell lines are differentially influenced. In addition, we examined two recently reported approaches of simultaneous quenching and extraction that showed minimal metabolome leakage. We also investigated the culture of cells on beads for rapid quenching and extraction, as a novel sample handling protocol. For metastatic breast cancer cells MDA-MB-436, the two direct quenching approaches and the bead harvesting approach showed favourable results with respect to metabolome leakage, compared to the conventional approaches. We characterised the recovery of eleven different classes of metabolites identified by gas chromatography–mass spectrometry in the cell extracts and the supernatants following quenching. Analysis of results based on metabolite classes is shown to be a useful approach aiding metabolomic interpretations. We also examined the effect of including a protein precipitation step on the metabolite classes detected. The de-proteinization step did not show significant improvement in overall recoveries. This investigation suggests that it is important to establish the level of metabolome leakage for the specific cell line investigated, irrespective of the methodology employed. Rapid approaches that combine quenching and extraction steps may be more effective in retaining valid metabolome data, with minimal metabolome leakage occurring.




Similar content being viewed by others
Abbreviations
- GC:
-
Gas chromatography
- MS:
-
Mass spectrometry
- GC–MS:
-
Gas chromatography–mass spectrometry
- LC–MS:
-
Liquid chromatography–mass spectrometry
- NMR:
-
Nuclear magnetic resonance spectroscopy
- TIC:
-
Total ion current
- PCA:
-
Principal component analysis
- AP:
-
Acetone precipitation
- NIST:
-
National Institute of Standards and Technology
- GMD:
-
Golm metabolome database
- AMDIS:
-
Automated mass spectral deconvolution and identification system
- ATCC:
-
American type culture collection
- Da:
-
Dalton
- EDTA:
-
Ethylene di-amine tetra-acetic acid
- MSTFA:
-
N-Methyl-N-(trimethylsilyl) trifluoroacetamide
- LN2:
-
Liquid nitrogen
- AMBIC:
-
Ammonium bicarbonate
- PBS:
-
Phosphate-buffered saline
References
Aranibar, N., Borys, M., Mackin, N. A., Ly, V., Abu-Absi, N., Abu-Absi, S., et al. (2011). NMR-based metabolomics of mammalian cell and tissue cultures. Journal of Biomolecular NMR, 49(3), 195–206.
Beloueche-Babari, M., Jackson, L. E., Al-Saffar, N. M. S., Eccles, S. A., Raynaud, F. I., Workman, P., et al. (2006). Identification of magnetic resonance detectable metabolic changes associated with inhibition of phosphoinositide 3-kinase signaling in human breast cancer cells. Molecular Cancer Therapeutics, 5(1), 187–196.
Bi, H., Krausz, K., Manna, S., Li, F., Johnson, C., & Gonzalez, F. (2013). Optimization of harvesting, extraction, and analytical protocols for UPLC-ESI-MS-based metabolomic analysis of adherent mammalian cancer cells. Analytical and Bioanalytical Chemistry, 405(15), 5279–5289. doi:10.1007/s00216-013-6927-9.
Bruce, S. J., Tavazzi, I., Parisod, V., Rezzi, S., Kochhar, S., & Guy, P. A. (2009). Investigation of human blood plasma sample preparation for performing metabolomics using ultrahigh performance liquid chromatography/mass spectrometry. Analytical Chemistry, 81(9), 3285–3296.
Dang, N.-H. T., Singla, A. K., Mackay, E. M., Jirik, F. R., & Weljie, A. M. (2014). Targeted cancer therapeutics: biosynthetic and energetic pathways characterized by metabolomics and the interplay with key cancer regulatory factors. Current Pharmaceutical Design, 20(15), 2637–2647.
Danielsson, A. P. H., Moritz, T., Mulder, H., & Spégel, P. (2010). Development and optimization of a metabolomic method for analysis of adherent cell cultures. Analytical Biochemistry, 404(1), 30–39.
Deja, S., Porebska, I., Kowal, A., Zabek, A., Barg, W., Pawelczyk, K., et al. (2014). Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. Journal of Pharmaceutical and Biomedical Analysis, 100, 369–380.
Dettmer, K., Nürnberger, N., Kaspar, H., Gruber, M. A., Almstetter, M. F., & Oefner, P. J. (2011). Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Analytical and Bioanalytical Chemistry, 399(3), 1127–1139.
Dietmair, S., Hodson, M. P., Quek, L. E., Timmins, N. E., Chrysanthopoulos, P., Jacob, S. S., et al. (2012). Metabolite profiling of CHO cells with different growth characteristics. Biotechnology and Bioengineering, 109, 1040–1414.
Dietmair, S., Timmins, N. E., Gray, P. P., Nielsen, L. K., & Krömer, J. O. (2010). Towards quantitative metabolomics of mammalian cells: Development of a metabolite extraction protocol. Analytical Biochemistry, 404(2), 155–164.
Duan, X., Young, R., Straubinger, R. M., Page, B. J., Cao, J., Wang, H., et al. (2009). A straightforward and highly efficient precipitation/on-pellet digestion procedure coupled to a long gradient nano-lc separation and orbitrap mass spectrometry for label-free expression profiling of the swine heart mitochondrial proteome. Journal of Proteome Research, 8(6), 2838.
Duarte, N. C., Becker, S. A., Jamshidi, N., Thiele, I., Mo, M. L., Vo, T. D., et al. (2007). Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proceedings of the National Academy of Sciences, 104(6), 1777–1782.
Dwivedi, P., Wu, P., Klopsch, S. J., Puzon, G. J., Xun, L., & Hill, H. H. (2008). Metabolic profiling by ion mobility mass spectrometry (IMMS). Metabolomics, 4(1), 63–80.
Fiehn, O. (2002). Metabolomics–the link between genotypes and phenotypes. Plant Molecular Biology, 48(1–2), 155–171.
Huang, G., Liu, X., Jiao, L., Xu, C., Zhang, Z., Wang, L., et al. (2014). Metabolomic evaluation of the response to endocrine therapy in patients with prostate cancer. European Journal of Pharmacology, 729, 132–137.
Hutschenreuther, A., Kiontke, A., Birkenmeier, G., & Birkemeyer, C. (2012). Comparison of extraction conditions and normalization approaches for cellular metabolomics of adherent growing cells with GCMS. Anal Methods, 4, 1953–1963.
Kronthaler, J., Gstraunthaler, G., & Heel, C. (2012). Optimizing high-throughput metabolomic biomarker screening: A study of quenching solutions to freeze intracellular metabolism in CHO cells. OMICS: A Journal of Integrative Biology, 16(3), 90–97.
Lane, A. N., & Fan, T. W. M. (2007). Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using H-1 TOCSY. Metabolomics, 3(2), 79–86. doi:10.1007/s11306-006-0047-x.
Lorenz, M. A., Burant, C. F., & Kennedy, R. T. (2011). Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical Chemistry, 83(9), 3406–3414.
Martano, G., Delmotte, N., Kiefer, P., Christen, P., Kentner, D., Bumann, D., et al. (2015). Fast sampling method for mammalian cell metabolic analyses using liquid chromatography–mass spectrometry. Nature Protocols, 10(1), 1–11.
Martineau, E., Tea, I., Loaëc, G., Giraudeau, P., & Akoka, S. (2011). Strategy for choosing extraction procedures for NMR-based metabolomic analysis of mammalian cells. Analytical and Bioanalytical Chemistry, 401(7), 2133–2142.
McDunn, J. E., Li, Z., Adam, K. P., Neri, B. P., Wolfert, R. L., Milburn, M. V., et al. (2013). Metabolomic signatures of aggressive prostate cancer. The Prostate, 73(14), 1547–1560.
Metallo, C. M., Walther, J. L., & Stephanopoulos, G. (2009). Evaluation of 13 C isotopic tracers for metabolic flux analysis in mammalian cells. Journal of Biotechnology, 144(3), 167–174.
Monton, M. R. N., & Soga, T. (2007). Metabolome analysis by capillary electrophoresis–mass spectrometry. Journal of Chromatography A, 1168(1), 237–246.
Schug, Z. T., Peck, B., Jones, D. T., Zhang, Q., Grosskurth, S., Alam, I. S., et al. (2015). Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell, 27(1), 57–71.
Sellick, C. A., Hansen, R., Stephens, G. M., Goodacre, R., & Dickson, A. J. (2011). Metabolite extraction from suspension-cultured mammalian cells for global metabolite profiling. Nature Protocols, 6(8), 1241–1249.
Sheikh, K. D., Khanna, S., Byers, S. W., Fornace, A. J, Jr, & Cheema, A. K. (2011). Small molecule metabolite extraction strategy for improving LC/MS detection of cancer cell metabolome. Journal of Biomolecular Techniques, 22(1), 1.
Stebbing, J., & Ellis, P. (2012). An overview of drug development for metastatic breast cancer. British Journal of Nursing, 21(Sup4), S18–S22.
Sumner, L. W., Mendes, P., & Dixon, R. A. (2003). Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry, 62(6), 817–836.
Teng, Q., Huang, W., Collette, T. W., Ekman, D. R., & Tan, C. (2009). A direct cell quenching method for cell-culture based metabolomics. Metabolomics, 5(2), 199–208.
Tiziani, S., Einwas, A.-H., Lodi, A., Ludwig, C., Bunce, C. M., Viant, M. R., et al. (2008). Optimized metabolite extraction from blood serum for H-1 nuclear magnetic resonance spectroscopy. Analytical Biochemistry, 377(1), 16–23. doi:10.1016/j.ab.2008.01.037.
Volmer, M., Gettmann, J., Scholz, S., Büntemeyer, H., & Noll, T. (2011). A method for metabolomic sampling of suspended animal cells using fast filtration. In BMC proceedings (Vol. 5, p. P93). London: BioMed Central Ltd.
Wiendahl, C., Brandner, J., Küppers, C., Luo, B., Schygulla, U., Noll, T., et al. (2007). A microstructure heat exchanger for quenching the metabolism of mammalian cells. Chemical Engineering and Technology, 30(3), 322–328.
Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., et al. (2008). Global metabolic profiling of Escherichia coli cultures: An evaluation of methods for quenching and extraction of intracellular metabolites. Analytical Chemistry, 80(8), 2939–2948. doi:10.1021/ac7023409.
Yuan, W., Anderson, K. W., Li, S., & Edwards, J. L. (2012). Subsecond absolute quantitation of amine metabolites using isobaric tags for discovery of pathway activation in mammalian cells. Analytical Chemistry, 84(6), 2892–2899.
Acknowledgments
Funding support from Chemical Engineering at the Life Science Interface (ChELSI) (EPSRC EP/E036252/1) for part funding of a PhD studentship to RK and the BBSRC DTA (BB/F016840/1) studentship funding to RC are gratefully acknowledged.
Author Contributions
Rahul Vijay Kapoore, Nicola J. Brown and Seetharaman Vaidyanathan conceived and designed the biological study. Rahul Vijay Kapoore and Seetharaman Vaidyanathan designed the metabolomics investigations. Rahul Vijay Kapoore and Rachael Coyle carried out the lab work. Rahul Vijay Kapoore analysed the metabolomics data. Rahul Vijay Kapoore wrote the manuscript with correction and input by all authors. Rachael Coyle contributed to biological experimentation. Seetharaman Vaidyanathan contributed to the statistical assessment of the data. All authors have given approval to the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kapoore, R.V., Coyle, R., Staton, C.A. et al. Cell line dependence of metabolite leakage in metabolome analyses of adherent normal and cancer cell lines. Metabolomics 11, 1743–1755 (2015). https://doi.org/10.1007/s11306-015-0833-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-015-0833-4