Abstract
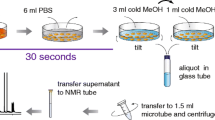
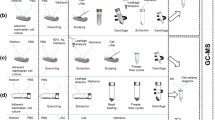
Trypsin/ethylenediaminetetraacetic acid (EDTA) treatment and cell scraping in a buffer solution were compared for harvesting adherently growing mammalian SW480 cells for metabolomics studies. In addition, direct scraping with a solvent was tested. Trypsinated and scraped cell pellets were extracted using seven different extraction protocols including pure methanol, methanol/water, pure acetone, acetone/water, methanol/chloroform/water, methanol/isopropanol/water, and acid–base methanol. The extracts were analyzed by GC-MS after methoximation/silylation and derivatization with propyl chloroformate, respectively. The metabolic fingerprints were compared and 25 selected metabolites including amino acids and intermediates of energy metabolism were quantitatively determined. Moreover, the influence of freeze/thaw cycles, ultrasonication and homogenization using ceramic beads on extraction yield was tested. Pure acetone yielded the lowest extraction efficiency while methanol, methanol/water, methanol/isopropanol/water, and acid–base methanol recovered similar metabolite amounts with good reproducibility. Based on overall performance, methanol/water was chosen as a suitable extraction solvent. Repeated freeze/thaw cycles, ultrasonication and homogenization did not improve overall metabolite yield of the methanol/water extraction. Trypsin/EDTA treatment caused substantial metabolite leakage proving it inadequate for metabolomics studies. Gentle scraping of the cells in a buffer solution and subsequent extraction with methanol/water resulted on average in a sevenfold lower recovery of quantified metabolites compared with direct scraping using methanol/water, making the latter one the method of choice to harvest and extract metabolites from adherently growing mammalian SW480 cells.






Similar content being viewed by others
References
Dettmer K, Aronov PA, Hammock BD (2007) Mass spectrometry-based metabolomics. Mass Spectrom Rev 26(1):51–78
Theobald U, Mailinger W, Reuss M, Rizzi M (1993) In vivo analysis of glucose-induced fast changes in yeast adenine nucleotide pool applying a rapid sampling technique. Anal Biochem 214(1):31–37
Villas-Boas SG, Hojer-Pedersen J, Akesson M, Smedsgaard J, Nielsen J (2005) Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast 22(14):1155–1169
Ritter JB, Genzel Y, Reichl U (2008) Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal Biochem 373(2):349–369
Bennett BD, Yuan J, Kimball EH, Rabinowitz JD (2008) Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3(8):1299–1311
Hofmann U, Maier K, Niebel A, Vacun G, Reuss M, Mauch K (2008) Identification of metabolic fluxes in hepatic cells from transient 13c-labeling experiments: part I. Exp Observations Biotechnol Bioeng 100(2):344–354
Teng Q, Huang WL, Collette TW, Ekman DR, Tan C (2009) A direct cell quenching method for cell-culture based metabolomics. Metabolomics 5(2):199–208
Maharjan RP, Ferenci T (2003) Global metabolite analysis: the influence of extraction methodology on metabolome profiles of escherichia coli. Anal Biochem 313(1):145–154
Timischl B, Dettmer K, Kaspar H, Thieme M, Oefner PJ (2008) Development of a quantitative, validated capillary electrophoresis-time of flight-mass spectrometry method with integrated high-confidence analyte identification for metabolomics. Electrophoresis 29(10):2203–2214
Hiller J, Franco-Lara E, Weuster-Botz D (2007) Metabolic profiling of escherichia coli cultivations: evaluation of extraction and metabolite analysis procedures. Biotechnol Lett 29(8):1169–1178
Winder CL, Dunn WB, Schuler S, Broadhurst D, Jarvis R, Stephens GM, Goodacre R (2008) Global metabolic profiling of escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal Chem 80(8):2939–2948
Rabinowitz JD, Kimball E (2007) Acidic acetonitrile for cellular metabolome extraction from escherichia coli. Anal Chem 79(16):6167–6173
Sellick C, Knight D, Croxford A, Maqsood A, Stephens G, Goodacre R, Dickson A (2010) Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: matching extraction approaches to cell type and metabolite targets. Metabolomics 6(3):427–438
Kaspar H, Dettmer K, Gronwald W, Oefner PJ (2008) Automated GC-MS analysis of free amino acids in biological fluids. J Chromatogr B Analyt Technol Biomed Life Sci 870(2):222–232
F.A.D.A. U. S. Department of Health and Human Services CfDEaR, Center for Veterinary Medicine (2001) Guidance for industry: bioanalytical method validation.
Klein MS, Almstetter MF, Schlamberger G, Nurnberger N, Dettmer K, Oefner PJ, Meyer HH, Wiedemann S, Gronwald W (2010) Nuclear magnetic resonance and mass spectrometry-based milk metabolomics in dairy cows during early and late lactation. J Dairy Sci 93(4):1539–1550
R-Development CoreTeam (2007) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Halket JM, Waterman D, Przyborowska AM, Patel RK, Fraser PD, Bramley PM (2005) Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J Exp Bot 56(410):219–243
Steinhauser D, Kopka J (2007) Methods, applications and concepts of metabolite profiling: primary metabolism. EXS 97:171–194
Acknowledgement
This study was funded in part by BayGene and the intramural ReForM C program of the Regensburg School of Medicine. The help of Dr. Y. Reinders, Regina Lindner, and Ruth Spaeth is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 10 kb)
Rights and permissions
About this article
Cite this article
Dettmer, K., Nürnberger, N., Kaspar, H. et al. Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols. Anal Bioanal Chem 399, 1127–1139 (2011). https://doi.org/10.1007/s00216-010-4425-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-4425-x