Abstract
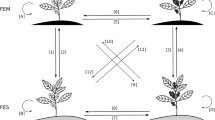
Genomic selection (GS) has been studied in several crops to increase the rates of genetic gain and reduce the length of breeding cycles. Despite its relevance, there are only a modest number of reports applied to the genus Coffea. Effective implementation depends on the ability to consider genomic models, which correctly represent breeding scenario in which the species are inserted. Coffee experimentation, in general, is represented by evaluations in multiple locations and harvests to understand the interaction and predict the performance of untested genotypes. Therefore, the main objective of this study was to investigate GS models suitable for use in Coffea canephora. An expansion of traditional GBLUP was considered and genomic analysis was performed using a genotyping-by-sequencing (GBS) approach, showed good potential to be used in coffee breeding programs. Interactions were modeled using the multiplicative mixed model theory, which is commonly used in multi-environment trials (MET) analysis in perennial crops. The effectiveness of the method used was compared with other genetic models in terms of goodness-of-fit statistics and prediction accuracy. Different scenarios that mimic coffee breeding were used in the cross-validation process. The method used had the lowest AIC and BIC values and, consequently, the best fit. In terms of predictive ability, the incorporation of the MET modeling showed higher accuracy (on average 10–17% higher) and lower prediction errors than traditional GBLUP. The results may be used as basis for additional studies into the genus Coffea and can be expanded for similar perennial crops.


Similar content being viewed by others
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19(6):716–723
Bates D, Vazquez AI (2014) pedigreemm: Pedigree-based mixed-effects models. https://CRAN.R-project.org/package=pedigreemm https://CRAN.R-project.org/package=pedigreemm, r package version 0.3-3
Burgueño J, Crossa J, Cotes JM, Vicente FS, Das B (2011) Prediction assessment of linear mixed models for multienvironment trials. Crop Sci 51(3):944–954
Burgueño J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52(2):707
de los Campos G, Naya H, Gianola D, Crossa J, Legarra A, Manfredi E, Weigel K, Cotes JM (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182(1):375–385
de los Campos G, Hickey JM, Pong-Wong R, Daetwyler HD, Calus MPL (2013) Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 193(2):327–45
Cilas C, Montagnon C, Bar-Hen A (2011) Yield stability in clones of coffea canephora in the short and medium term: longitudinal data analyses and measures of stability over time. Tree Genet Genomes 7(2):421–429
Crossa J, Burgueño J, Cornelius PL, McLaren G, Trethowan R, Krishnamachari A (2006) Modeling genotype × environment interaction using additive genetic covariances of relatives for predicting breeding values of wheat genotypes. Crop Sci 46(4):1722–1733
Crossa J, Beyene Y, Kassa S, Pérez P, Hickey JM, Chen C, de los Campos G, Burgueño J, Windhausen VS, Buckler E et al (2013) Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3: Genes–Genomes–Genetics 3(11):1903–1926
Cubry P, De Bellis F, Avia K, Bouchet S, Pot D, Dufour M, Legnate H, Leroy T (2013) An initial assessment of linkage disequilibrium (ld) in coffee trees: Ld patterns in groups of coffea canephora pierre using microsatellite analysis. BMC Genomics 14 (1):10
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST et al (2011) The variant call format and vcftools. Bioinformatics 27(15):2156–2158
Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G et al (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345(6201):1181–1184
Elshire RJ, Glaubitz JC, Sun Q, Poland Ja, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6(5):e19,379
Falconer DS, Mackay TFC (1996) Quantitative genetics. Pearson Education Limited, England
Ferrão LFV, Caixeta ET, Souza FdF, Zambolim EM, Cruz CD, Zambolim L, Sakiyama NS (2013) Comparative study of different molecular markers for classifying and establishing genetic relationships in coffea canephora. Plant Syst Evol 299(1):225–238
Ferrão LFV, Caixeta ET, Pena G, Zambolim EM, Cruz CD, Zambolim L, Ferrão MAG, Sakiyama NS (2015) New EST–SSR markers of Coffea arabica: transferability and application to studies of molecular characterization and genetic mapping. Mol Breed 35(1):1–5
Ferrão LFV, Ferrão RG, Ferrão MAG, Fonseca A, Stephens M, Garcia AAF (2016) Genomic prediction in Coffea canephora using Bayesian polygenic modeling. In: 5th international conference on quantitative genetics. WI, Madison, p 203
Ferrão RG, Ferrão MAG, Fonseca A, Pacova B (2007) Melhoramento genético de Coffea canephora. In: Ferrão R, Fonseca A, Bragança S, Ferrão M, Muner LD (eds) Cafe conilon, incaper edn Vitória-ES, pp 123–173
Gelman A, Carlin JB, Stern HS, Rubin DB (2014) Bayesian data analysis, vol 2. Chapman & hall/CRC Boca Raton, FL, USA
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) Tassel-gbs: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9(2):e90,346
GNU P (2007) Free Software Foundation. Bash (3.2.48) [Unix shell program].Retrieved from http://ftp.gnu.org/gnu/bash/bash-3.2.48.tar.gz
Grattapaglia D, Resende MDV (2010) Genomic selection in forest tree breeding. Tree Genet Genomes 7(2):241–255
Heslot N, Akdemir D, Sorrells ME, Jannink JL (2014) Integrating environmental covariates and crop modeling into the genomic selection framework to predict genotype by environment interactions. Theor Appl Genet 127(2):463–480
Hu Y, Yan C, Hsu CH, Chen QR, Niu K, Komatsoulis GA, Meerzaman D (2014) Omiccircos: a simple-to-use r package for the circular visualization of multidimensional omics data. Cancer Informat 13:13
IOC (2016) International Coffee Organization - Trade Statistics Tables. http://www.ico.org/
Jannink JL, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Brief Funct Genomics 9(2):166–177
Jarquín D, Crossa J, Lacaze X, Du Cheyron P, Daucourt J, Lorgeou J, Piraux F, Guerreiro L, Pérez P, Calus M et al (2014a) A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor Appl Genet 127(3):595–607
Jarquín D, Kocak K, Posadas L, Hyma K, Jedlicka J, Graef G, Lorenz A (2014b) Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genomics 15(1):740
Kelly AM, Cullis BR, Gilmour AR, Eccleston JA, Thompson R (2009) Estimation in a multiplicative mixed model involving a genetic relationship matrix. Genet Sel Evol 41(1):1
Lopez-Cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink JL, Singh RP, Autrique E, de los Campos G (2015) Increased prediction accuracy in wheat breeding trials using a marker x environment interaction genomic selection model. G3: Genes–Genomes–Genetics 5(4):569–82
Malosetti M, Ribaut JM, van Eeuwijk FA (2014) The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Drought phenotyping in crops: From theory to practice 4(44):53
Malosetti M, Bustos-Korts D, Boer MP, van Eeuwijk FA (2016) Predicting responses in multiple environments: issues in relation to genotype × environment interactions. Crop Sci 56(5):2210–2222
Margarido GRA, Pastina MM, Souza AP, Garcia AAF (2015) Multi-trait multi-environment quantitative trait loci mapping for a sugarcane commercial cross provides insights on the inheritance of important traits. Mol Breed 35(8):175
Mérot-L’Anthoëne V, Mangin B, Lefebvre-Pautigny F, Jasson S, Rigoreau M, Husson J, Lambot C, Crouzillat D (2014) Comparison of three qtl detection models on biochemical, sensory, and yield characters in coffea canephora. Tree Genet Genomes 10(6):1541–1553
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Meyer K (2009) Factor-analytic models for genotype × environment type problems and structured covariance matrices. Genet Sel Evol 41(1):21
Mrode RA (2014) Linear models for the prediction of animal breeding values. Cabi
Oakey H, Cullis B, Thompson R, Comadran J, Halpin C, Waugh R (2016) Genomic selection in multi-environment crop trials. G3: Genes–Genomes–Genetics 6(5):1313–1326
Pastina MM, Malosetti M, Gazaffi R, Mollinari M, Margarido GRA, Oliveira KM, Pinto LR, Souza AP, van Eeuwijk FA, Garcia AAF (2012) A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Theor Appl Genet 124:835–849
Payne RW, Murray DA, Harding SA (2011) An introduction to the genstat command language (14th edn)
Piepho H, Möhring J, Melchinger A, Büchse A (2008) Blup for phenotypic selection in plant breeding and variety testing. Euphytica 161(1-2):209–228
Poland J, Endelman J, Dawson J, Rutkoski J, Wu S, Manes Y, Dreisigacker S, Crossa J, Sánchez-Villeda H, Sorrells M, Jannink JL (2012) Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome J 5(3):103
R Core Team (2013) R: A Language and Environment for Statistical Computing
Schulz-Streeck T, Ogutu JO, Piepho HP (2013) Comparisons of single-stage and two-stage approaches to genomic selection. Theor Appl Genet 126(1):69–82
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Smith AB, Cullis BR, Thompson R (2001) Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 57(4):1138–1147
Smith AB, Cullis BR, Thompson R (2005) The analysis of crop cultivar breeding and evaluation trials: an overview of current mixed model approaches. J Agric Sci 143(06):449
Smith AB, Stringer JK, Wei X, Cullis BR (2007) Varietal selection for perennial crops where data relate to multiple harvests from a series of field trials. Euphytica 157(1-2):253–266
Smith KF, Casler M (2004) Spatial analysis of forage grass trials across locations, years, and harvests. Crop Sci 44(1):56–62
Spindel JE, Begum H, Akdemir D, Collard B, Redona E, Jannink Jl, Mccouch S (2016) Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity 116:395–408
Tran HTM, Lee LS, Furtado A, Smyth H, Henry RJ (2016) Advances in genomics for the improvement of quality in coffee. J Sci Food Agric 96(10):3300–3312
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91(11):4414–4423
Wang W, Gelman A (2014) Difficulty of selecting among multilevel models using predictive accuracy. Statistics at its Interface 7(1):1–88
Acknowledgments
This work is partially supported by FAPESP/CAPES (São Paulo Research Foundation), grants 2014/20389-2 for L.F.V.F and A.A.F.G. Phenotypic evaluations and GBS data is supported by Fapes (Espírito Santo Research Foundation), grants 55207464/11 and 65192036/14. Additional support is provided by the Instituto Capixaba de Pesquisa, Assitência Tecnica e Extensão Rural (Incaper) and Embrapa Cafe. A.A.F.G, R.G.F, M.A.G.F and A.F have a fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The author thank Livia Souza and Anete P. de Souza (CBMEG, Unicamp/Brazil) by the assistance in the DNA extraction step; and Paulo Volpi (Incaper/Brazil) by the support on the phenotypic evaluation.
Author information
Authors and Affiliations
Contributions
L.F.V.F, A.A.F.G, R.G.F, M.A.G.F and A.F conceived the study and designed the experiments. R.G.F, M.A.G.F and A.F installed the experimental design and collected the phenotypic data. L.F.V.F performed the DNA extraction. L.F.V.F and A.A.F.G performed the genomic prediction analysis and proposed the theoretical idea of the model. L.F.V.F wrote the paper.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Data Archiving Statement
The genotypes in the study belong to the germplasm collection and breeding program of the Incaper institution (ES,Brazil). Phenotypic and genotypic data were submitted as Supplementary Material.
Additional information
Communicated by: J. Beaulieu
This article is part of the Topical Collection on Breeding
Key Message: First insights into the Genotyping-by-Sequencing (GBS) in Coffea canephora and a genomic prediction model considering the theory about multiplicative mixed model to accommodate the interaction effects across sites and harvests
Rights and permissions
About this article
Cite this article
Ventorim Ferrão, L., Gava Ferrão, R., Ferrão, M. et al. A mixed model to multiple harvest-location trials applied to genomic prediction in Coffea canephora . Tree Genetics & Genomes 13, 95 (2017). https://doi.org/10.1007/s11295-017-1171-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1171-7