Abstract
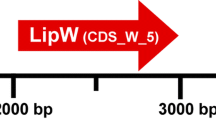
In order to search new lipolytic enzymes and conduct bioprospecting of microbial communities from high Andean forest soil, a metagenomic library of approximately 20,000 clones was constructed in Escherichia coli using plasmid p-Bluescript II SK+. The library covered 80 Mb of the metagenomic DNA mainly from Proteobacteria, Actinobacteria and Acidobacteria. Two clones with lipolytic activity in tributyrin as a substrate were recovered. Clone BAA3G2 (pSK-estGX1) was selected and the entire 4.6 Kb insert sequence was determined. The sequence had a GC content of 70.6% and could be derived from an undescribed Actinobacteria genome. One open reading frame encoded a polypeptide of 210 amino acids (gene estGX1) with a molecular mass of 22.4 kDa that contained the pentapeptide G-P-S-G-G near the N-terminus essential for lipase activity and the putative catalytic triad was identified, also a putative ribosomal binding site located 18 bp upstream the estGX1 ATG start codon was identified. The phylogenetic analysis suggested that the protein belonged to a new lipase family. The secreted enzyme showed a preference for short length fatty acids, with specific activity against p-nitrophenyl-butyrate (0.142 U/mg of total protein), it was cold active with relative activity of 30% at 10°C and moderately thermo active with relative activity of 80% at 50°C and had a pH optimum of 8.0 at 40°C.






Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59(1):143–169
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343:177–183
Bornscheuer U (2002) Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev 26:73–81
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bunterngsook B, Kanokratana P, Thongaram T, Tanapongpipat S, Uengwetwanit T, Rachdawong S, Vichitsoonthonkul T, Eurwilaichitr L (2010) Identification and characterization of lipolityc enzymes froma a Peat-swamp forest soil metagenome. Biosci Biotecnol Biochem 74(9):1848–1854. doi:10.1271/bbb.100249
Chao W, Sun B (2009) Identification of novel esterase from metagenomic library of Yangtze River. J Microbiol Biotechnol 19(2):187–193. doi:10.4014/jmb.0804.292
Chu X, He H, Guo C, Sun Baolin (2008) Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol 80:615–625. doi:10.1007/s00253-008-1566-3
Côté A, Shareck F (2010) Expression and characterization of a novel heterologous moderately thermostable lipase derived from metagenomics in Streptomyces lividans. J Ind Microbiol Biotechnol. doi:10.1007/s10295-010-0735-4
Couto GH, Glogauer A, Faoro A, Chubatsu LS, Souza EM, Pedrosa FO (2010) Isolation of a novel lipase from a metagenomic library derived from mangrove sediment from the south Brazilian coast. Genet Mol Res 9(1):514–523
Daniel R (2005) The metagenomics of soil. Nat Rev Microbiol 3:470–478
Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD et al (2006) Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol 72(5):3637–3645. doi:10.1128/Aem.72.5.3637-3645.2006
Elend C, Schmeisser C, Hoebenreich H, Steele HL, Streit WR (2007) Isolation and characterization of a metagenome-derived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J Biotechnol 130:370–377. doi:10.1016/j.jbiotec.2007.05.015
Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, Santos VA, Strompl C, Elborough K, Jarvis G, Neef A, Yakimov MM, Timmis KN, Golyshin PN (2005) Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol 7(12):1996–2010. doi:10.1111/j.1462-2920.2005.00920.x
Fischer M, Pleiss J (2003) The lipase engineering database: a navigation and analysis tool for protein families. Nucleic Acids Res 31(1):319–321. doi:10.1093/nar/gkg015
Gabor EM, Alkema WB, Janssen DB (2004) Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 6:879–886. doi:10.1111/j.1462-2920.2004.00640.x
Handelsman J et al (2007) The new science of metagenomics: revealing the secrets of our microbial planet. The National Academies Press, Washington
Hårdeman F, Sjöling S (2006) Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol 59:524–534. doi:10.1111/j.1574-6941.2006.00206.x
Heath C, Hu XP, Cary CS, Cowan D (2009) Isolation and characterisation of a novel, low-temperature-active alkaliphilic esterase from an Antarctic desert soil metagenome. Appl Environ Microbiol 75(13):4657–4659. doi:10.1128/AEM.02597-08
Henne A, Schmitz RA, Bömeke M, Gottschalk G, Daniel R (2000) Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity of Escherichia coli. Appl Environ Microbiol 66(7):3113–3116
Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A (2004) ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res 32:145–147. doi:10.1093/nar/gkh141
Houde A, Kademi A, Leblanc D (2004) Lipases and their industrial applications: an overview. Appl Biochem Biotechnol 118:155–170
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Jaeger KE, Reetz MT (1998) Microbial lipases form versatile tools for biotechnology. Trends Biotechnol 16(9):396–403. doi:10.1016/S0167-7799(98)01195-0
Jaeger KE, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53:315–351
Jeon JH, Kim JT, Kim YJ, Kim HK, Lee HS, Kang SG, Kim SJ, Lee JH (2009) Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl Microbiol Biotechnol 81:865–874. doi:10.1007/s00253-008-1656-2
Kim EY, Oh KH, Lee MH, Kang CH, Oh TK, Yoon JH (2009a) Novel cold-adapted alkaline lipase from intertidal flat metagenome and proposal of a new family of bacterial lipase. Appl Environ Microbiol 75(1):257–260. doi:10.1128/AEM.01400-08
Kim JS, Lim HK, Lee MH, Park JH, Hwang EC, Moon BJ, Lee SW (2009b) Production of porphyrin intermediates in Escherichia coli carrying soil metagenomic genes. FEMS Microbiol Lett 295:42–49. doi:10.1111/j.1574-6968.2009.01577.x
Kim YH, Kwon EJ, Kim SK, Jeong YS, Kim J, Yun HD, Kim H (2010) Molecular cloning and characterization of a novel family VIII alkaline esterase from a compost metagenomic library. Biochem Biophys Res Commun 393:45–49. doi:10.1016/j.bbrc.2010.01.070
Kumar S, Dudley J, Nei M, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306. doi:10.1093/bib/bbn017
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi:10.1038/227680a0
Lämmle K, Zipper H, Breuer M, Hauer B, Buta C, Brunner H, Rupp S (2007) Identification of novel enzymes with different hydrolytic activities by metagenome expression cloning. J Biotechnol 127:575–592. doi:10.1016/j.jbiotec.2006.07.036
Lane LC (1978) A simple method for stabilizing protein sulfhydryl groups during SDS-gel electrophoresis. Anal Biochem 86(2):655–665. doi:10.1016/0003-2697(78)90792-3
Lee SW, Won K, Lim HK, Kim JC, Choi GJ, Cho KY (2004) Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl Microbiol Biotechnol 65:720–726. doi:10.1007/s00253-004-1722-3
Lee MH, Lee CH, Oh TK, Song JK, Yoon JH (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72(11):7406–7409. doi:10.1128/Aem.01157-06
Lee MH, Hong KS, Malhotra S, Park JH, Hwang EC, Choi HK, Kim YS, Tao W, Lee SW (2010) A new esterase EstD2 isolated from plant rhizosphere soil metagenome. Appl Microbiol Biotechnol. doi:10.1007/s00253-010-2729-6
Li G, Wang K, Liu Y (2008) Molecular cloning and characterization of a novel pyrethroid-hydrolyzing esterase originating from the Metagenome. Microb Cell Fact 7:38. doi:10.1186/1475-2859-7-38
Liaw RB, Cheng MP, Wu MC, Lee CY (2010) Use of metagenomic approaches to isolate lipolytic genes from activated sludge. Bioresour Technol 101:8323–8329. doi:10.1016/j.biortech.2010.05.091
Liu K, Wang J, Bu D, Zhao S, McSweeney C, Yu P, Li D (2009) Isolation and biochemical characterization of two lipases from a metagenomic library of China Holstein cow rumen. Biochem and Biophys Res Commun 385:605–611. doi:10.1016/j.bbrc.2009.05.110
Martinez A, Kolvek SJ, Tiong Yip CLT, Hopke J, Brown KA, MacNeil IA, Osburne MS (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70(4):2452–2463. doi:10.1128/AEM.70.4.2452-2463.2004
Meilleur C, Hupe JF, Juteau P, Shareck F (2009) Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenomic library. J Ind Microbiol Biotechnol 36(6):853–861. doi:10.1007/s10295-009-0562-7
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9(386):1–8. doi:10.1186/1471-2105-9-386
Misawa E, Chion CK, Archer IV, Woodland MP, Zhou NY, Carter SF, Widdowson DA, Leak DJ (1998) Characterization of a catabolic epoxide hydrolase from a Corynebacterium sp. Eur J Biochem 253(1):173–183. doi:10.1046/j.1432-1327.1998.2530173.x
Rajan M (2004) Global market for industrial enzymes to reach $2.4 billion by 2009. Business Communications Company, Inc. RC-147U enzymes for industrial applications. http://www.bccresearch.com/editors/RC147U.html
Ranjan R, Grover A, Kapardar RK, Sharma R (2005) Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem Biophys Res Commun 335:57–65. doi:10.1016/j.bbrc.2005.07.046
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schepens H (2003) White biotechnology: gateway to a more sustainable future [online] http://www.mckinsey.com/clientservice/chemicals/pdf/BioVision_Booklet_final.pdf. Accessed 15 Sep 2010
Schmeisser C, Steele H, Streit WR (2007) Metagenomics, biotechnology with non-culturable microbes. Appl Microbiol Biotechnol 75(5):955–962. doi:10.1007/s00253-007-0945-5
Schnoes AM, Brown SD, Babbitt PC (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5(12):e1000605. doi:10.1371/journal.pcbi.1000605
Shu ZY, Jiang H, Lin RF, Jiang YM, Lin L, Huang JZ (2010) Technical methods to improve yield, activity and stability in the development of microbial lipases. J Mol Catal B: Enzym 62:1–8. doi:10.1016/j.molcatb.2009.09.003
Sik HK, Lim HK, Chung EJ, Park EJ, Lee MH, Kim JC, Choi GJ, Cho KY, Lee SW (2007) Selection and characterization of forest soil metagenome genes encoding lipolytic enzymes. J Microbiol Biotechnol 17(10):1655–1660
Simon C, Daniel R (2009) Achievements and new knowledge unraveled by metagenomic approaches. Appl Microbiol Biotechnol 85:265–276. doi:10.1007/s00253-009-2233-z
Sjöling S, Sttaford W, Cowan D (2007) Soil metagenomic: exploring and exploting the soil Microbial Gen Pool. In: Van Elsas JD, Janson J, Trevors J (eds) Modern soil microbiology. CRC Press, New York, pp 409–434
Steele HE, Jaeger JE, Daniel R, Streit WR (2009) Advances in recovery of novel biocatalysts from metagenomes. J Mol Microbiol Biotechnol 16(1–2):25–37. doi:10.1159/000142892
Terahara T, Yamada K, Kurata S, Yokomaku T, Tsuneda S, Harayama S (2010) Direct cloning and expression of putative esterase genes from environmental DNA. Enzyme Microb Technol 47:17–23. doi:10.1016/j.enzmictec.2010.04.005
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tirawongsaroj P, Sriprang R, Harnpicharnchai P, Thongarama T, Champreda V, Tanapongpipat S, Pootanakit K, Eurwilaichitr L (2008) Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J Biotechnol 133:42–49. doi:10.1016/j.jbiotec.2007.08.046
Uchiyama T, Miyazaki K (2009) Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol 20:616–622. doi:10.1016/j.copbio.2009.09.010
Wei P, Bai L, Song W, Hao G (2009) Characterization of two soil metagenome-derived lipases with high specificity for p-nitrophenyl palmitate. Arch Microbiol 191:233–240. doi:10.1007/s00203-008-0448-5
Zhang T, Han WJ, Liu ZP (2009) Gene cloning and characterization of a novel esterase from activated sludge metagenome. Microb Cell Fact 8:67. doi:10.1186/1475-2859-8-67
Acknowledgments
We thank I. Balzer for useful discussion, M. M. Zambrano for reviewing the manuscript and D. Borda for library screening. This research was supported by Department of Science, Technology and Innovation (COLCIENCIAS)—SENA (project number 6570-392-19990) and was done under MAVDT (contract number 15, 2008) for access to genetic resources and UAESPNN (research permit code DTNO-N-20/2007).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiménez, D.J., Montaña, J.S., Álvarez, D. et al. A novel cold active esterase derived from Colombian high Andean forest soil metagenome. World J Microbiol Biotechnol 28, 361–370 (2012). https://doi.org/10.1007/s11274-011-0828-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0828-x