Abstract
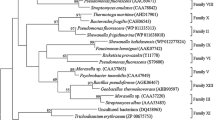
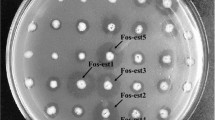
The demand for novel biocatalysts is increasing in modern biotechnology, which greatly stimulates the development of powerful tools to explore the genetic resources in the environment. Metagenomics, a culture independent strategy, provides an access to valuable genetic resources of the uncultured microbes. In this study, two novel esterase genes designated as estA and estB, which encoded 277- and 328-amino-acid peptides, respectively, were isolated from a marine microbial metagenomic library by functional screening, and the corresponding esterases EstA and EstB were biochemically characterized. Amino acid sequence comparison and phylogenetic analysis indicated that EstA together with other putative lipolytic enzymes was closely related to family III, and EstB with its relatives formed a subfamily of family IV. Site-directed mutagenesis showed that EstA contained classical catalytic triad made up of S146–D222–H255, whereas EstB contained an unusual catalytic triad which consisted of S–E–H, an important feature of the subfamily. EstA exhibited habitat-specific characteristics such as its high level of stability in the presence of various divalent cations and at high concentrations of NaCl. EstB displayed remarkable activity against p-nitrophenyl esters and was highly stable in 30% methanol, ethanol, dimethylformamide, and dimethyl sulfoxide, making EstB a potential candidate for industrial applications.




Similar content being viewed by others
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343(Pt 1):177–183
Bell PJ, Sunna A, Gibbs MD, Curach NC, Nevalainen H, Bergquist PL (2002) Prospecting for novel lipase genes using PCR. Microbiology 148:2283–2291
Choo DW, Kurihara T, Suzuki T, Soda K, Esaki N (1998) A cold-adapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: gene cloning and enzyme purification and characterization. Appl Environ Microbiol 64:486–491
De Simone G, Galdiero S, Manco G, Lang D, Rossi M, Pedone C (2000) A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase. J Mol Biol 303:761–771
De Simone G, Menchise V, Manco G, Mandrich L, Sorrentino N, Lang D, Rossi M, Pedone C (2001) The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J Mol Biol 314:507–518
Elend C, Schmeisser C, Leggewie C, Babiak P, Carballeira JD, Steele HL, Reymond JL, Jaeger KE, Streit WR (2006) Isolation and biochemical characterization of two novel metagenome-derived esterases. Appl Environ Microbiol 72:3637–3645
Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Martins Dos Santos VA, Yakimov MM, Timmis KN, Golyshin PN (2005) Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chem Biol 12:895–904
Gupta R, Gupta N, Rathi P (2004) Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol 64:763–781
Hardeman F, Sjoling S (2007) Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol 59:524–534
Henne A, Daniel R, Schmitz RA, Gottschalk G (1999) Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol 65:3901–3907
Henne A, Schmitz RA, Bomeke M, Gottschalk G, Daniel R (2000) Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl Environ Microbiol 66:3113–3116
Jaeger KE, Reetz MT (1998) Microbial lipases form versatile tools for biotechnology. Trends Biotechnol 16:396–403
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Kurose K, Isozaki E, Tohkin M, Fukuhara M (1999) Cloning and expression analysis of a new member of the cytochrome P450, CYP2A15 from the Chinese hamster, encoding testosterone 7alpha-hydroxylase. Arch Biochem Biophys 371:270–276
Lee SW, Won K, Lim HK, Kim JC, Choi GJ, Cho KY (2004) Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl Microbiol Biotechnol 65:720–726
Lee MH, Lee CH, Oh TK, Song JK, Yoon JH (2006) Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Appl Environ Microbiol 72:7406–7409
Li X, Qin L (2005) Metagenomics-based drug discovery and marine microbial diversity. Trends Biotechnol 23:539–543
MacNeil IA, Tiong CL, Minor C, August PR, Grossman TH, Loiacono KA, Lynch BA, Phillips T, Narula S, Sundaramoorthi R, Tyler A, Aldredge T, Long H, Gilman M, Holt D, Osburne MS (2001) Expression and isolation of antimicrobial small molecules from soil DNA libraries. J Mol Microbiol Biotechnol 3:301–308
Manco G, Adinolfi E, Pisani FM, Ottolina G, Carrea G, Rossi M (1998) Overexpression and properties of a new thermophilic and thermostable esterase from Bacillus acidocaldarius with sequence similarity to hormone-sensitive lipase subfamily. Biochem J 332:203–212
Prim N, Bofill C, Pastor FI, Diaz P (2006) Esterase EstA6 from Pseudomonas sp. CR-611 is a novel member in the utmost conserved cluster of family VI bacterial lipolytic enzymes. Biochimie 88:859–867
Ranjan R, Grover A, Kapardar RK, Sharma R (2005) Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem Biophys Res Commun 335:57–65
Rees HC, Grant S, Jones B, Grant WD, Heaphy S (2003) Detecting cellulase and esterase enzyme activities encoded by novel genes present in environmental DNA libraries. Extremophiles 7:415–421
Rondon MR, Raffel SJ, Goodman RM, Handelsman J (1999) Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc Natl Acad Sci U S A 96:6451–6455
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schmeisser C, Stockigt C, Raasch C, Wingender J, Timmis KN, Wenderoth DF, Flemming HC, Liesegang H, Schmitz RA, Jaeger KE, Streit WR (2003) Metagenome survey of biofilms in drinking-water networks. Appl Environ Microbiol 69:7298–7309
Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF (1996) Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol 178:591–599
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Walter J, Mangold M, Tannock GW (2005) Construction, analysis, and beta-glucanase screening of a bacterial artificial chromosome library from the large-bowel microbiota of mice. Appl Environ Microbiol 71:2347–2354
Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J (2000) Novel natural products from soil DNA libraries in a streptomycete host. Org Lett 2:2401–2404
Wei Y, Contreras JA, Sheffield P, Osterlund T, Derewenda U, Kneusel RE, Matern U, Holm C, Derewenda ZS (1999) Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat Struct Biol 6:340–345
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, Jaroszewski L, Cieplak P, Miller CS, Li H, Mashiyama ST, Joachimiak MP, van Belle C, Chandonia JM, Soergel DA, Zhai Y, Natarajan K, Lee S, Raphael BJ, Bafna V, Friedman R, Brenner SE, Godzik A, Eisenberg D, Dixon JE, Taylor SS, Strausberg RL, Frazier M, Venter JC (2007) The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol 5:e16
Acknowledgments
This work was funded by the National High-Tech Research and Development Program of China (2007AA09Z421) and the One Hundred Talent Project of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chu, X., He, H., Guo, C. et al. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl Microbiol Biotechnol 80, 615–625 (2008). https://doi.org/10.1007/s00253-008-1566-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1566-3