Abstract
Aspergillus niger NCIM 563 produced two different extracellular phytases (Phy I and Phy II) under submerged fermentation conditions at 30°C in medium containing dextrin-glucose-sodium nitrate-salts. Both the enzymes were purified to homogeneity using Rotavapor concentration, Phenyl-Sepharose column chromatography and Sephacryl S-200 gel filtration. The molecular mass of Phy I and II as determined by SDS–PAGE and gel filtration were 66, 264, 150 and 148 kDa respectively, indicating that Phy I consists of four identical subunits and Phy II is a monomer. The pI values of Phy I and II were 3.55 and 3.91, respectively. Phy I was highly acidic with optimum pH of 2.5 and was stable over a broad pH range (1.5–9.0) while Phy II showed a pH optimum of 5.0 with stability in the range of pH 3.5–9.0. Phy I exhibited very broad substrate specificity while Phy II was more specific for sodium phytate. Similarly Phy II was strongly inhibited by Ag+, Hg2+ (1 mM) metal ions and Phy I was partially inhibited. Peptide analysis by Mass Spectrometry (MS) MALDI-TOF also indicated that both the proteins were totally different. The K m for Phy I and II for sodium phytate was 2.01 and 0.145 mM while V max was 5,018 and 1,671 μmol min−1 mg−1, respectively. The N-terminal amino acid sequences of Phy I and Phy II were FSYGAAIPQQ and GVDERFPYTG, respectively. Phy II showed no homology with Phy I and any other known phytases from the literature suggesting its unique nature. This, according to us, is the first report of two distinct novel phytases from Aspergillus niger.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytase, a phosphomonoesterase, is an enzyme capable of hydrolysing phytate [myo-inositol (1,2,3,4,5,6) hexakisphosphate], the major storage form of phosphorus in cereals and legumes, representing 18–88% of total phosphorus content (Konietzny and Greiner 2002). Phytases (EC 3.1.3.8 and EC 3.1.3.26) belong to the family of histidine acid phosphatases which hydrolyses phytate to liberate inositol and inorganic phosphate (Mullaney et al. 2000). Phytate phosphorus is biologically unavailable to non-ruminants as they lack or secrete a very low level of phytase activity in the intestine. Consequently, the phytate in animal feeds is discharged in the feces of these animals into rivers and seas, resulting in severe pollution of water resources (Mullaney et al. 2000). Similarly, phytic acid is an anti-nutrient, which due to its strong chelating potential can bind essential minerals such as calcium, zinc and copper, rendering them unavailable or poorly available for absorption (Singh 2008). To overcome this difficulty the feed has to be supplemented with inorganic phosphate to meet the nutritional requirements of the animals (Selle and Ravindran 2007). Supplementation of feed with phytases in contrast increases the bioavailability of phytic acid-bound phosphate, facilitating a reduction in the addition of inorganic phosphate to the feed and a decrease in phosphorus excretion in areas of intensive animal husbandry.
Although a large number of bacteria, yeasts and fungi are reported to produce phytase (Vohra and Satyanarayana 2003; Vats and Banerjee 2004; Kaur et al. 2007) fungal phytases are preferred in animal feed due to their high yield and more acid tolerance as compared to phytases from bacteria (Kim et al. 1998). Similarly the pH in poultry gut varies from 2.5 to 6.0, thus phytase active and stable in acidic environment is highly preferred (Radcliffe et al. 1998). Among fungi many Aspergilli (Ullah 1988; Vohra and Satyanarayana 2003; Vats and Banerjee 2004; Vats et al. 2004) are known to be active phytase producers. As Aspergillus niger is Generally Recognised as Safe (GRAS) it is frequently used in food and feed applications. Earlier we have reported phytase production by Aspergillus niger NCIM 563 under submerged fermentation (Soni and Khire 2007; Bhavsar et al. 2008; Shah et al. 2009) which includes production and partial characterization of two types of phytase from Aspergillus niger NCIM 563. The present communication reports purification and characterization of two novel phytases (Phy I and Phy II), which according to us, is the first report of two distinct forms of extracellular acidic phytases produced simultaneously under submerged fermentation.
Materials and methods
Chemicals
Phytic acid sodium salt was purchased from Sigma Chemical Company, St Louise, MO, USA. All other chemicals used were of analytical grade and obtained from leading manufacturers including BDH, Sigma and Glaxo. SDS–PAGE and gel filtration markers, Coomassie Brilliant Blue R-250 and Bromophenol Blue were purchased from Sigma Chemical Company, USA Sephacryl S-300, Phenyl-Sepharose CL-4B were obtained from Sigma.
Organism and culture conditions
The strain used throughout the present work was Aspergillus niger NCIM 563 (Soni and Khire 2007). It was maintained on Potato Dextrose Agar (PDA) slants. The fungus was grown in modified fermentation medium containing (per 100 ml): Dextrin 5 g; Glucose 2.5 g; NaNO3 0.86 g; KH2PO4 0.004 g; KCl 0.05 g; MgSO4·7H2O 0.05 g; FeSO4·7H2O 0.01 g. pH 5.5 before sterilization. Fermentation medium (100 ml in 250-ml Erlenmeyer flasks) was inoculated with 1% (v/v) of spore suspension (5 × 107 spores per ml) prepared by suspending the spores from 7-day-old sporulated slants of Aspergillus niger NCIM 563 grown on PDA in 10 ml of sterile distilled water containing 0.01% (v/v) Tween 80 and incubated at 30°C at 200 rev/min. Samples were removed after every 24 h and checked for pH, growth, total residual reducing sugar, extracellular protein and phytase activity.
Assay of phytase and protein
Phytase activity was measured at 55°C as described earlier (Soni and Khire 2007). The reaction for Phy I and Phy II was carried out at pH 2.5 (100 mM Glycine-HCl buffer) and pH 5.0 (100 mM acetate buffer) at 55°C for 30 min, respectively. The liberated inorganic phosphate was measured by a modification of the ammonium molybdate method (Heinohen and Lathi 1981). A freshly prepared 4 ml solution of acetone:2.5 M H2SO4:10 mM ammonium molybdate (2:1:1 v/v/v) and 400 μl of 1 M citric acid were added to the assay mixture. Absorbance was measured at 370 nm. One unit of phytase activity (IU) was expressed as the amount of enzyme that liberates 1 μmol phosphorus per minute under standard assay conditions.
Each experiment was carried out in triplicate and the values reported are the mean of three such experiments in which a maximum of 3–5% variability was observed.
Protein concentration in the fermentation broth and in the purified enzyme preparation was determined by the Lowry method as well as measurement of absorbance at 280 nm using BSA as a standard.
Purification of phytase
After fermentation, mycelium was separated by filtration followed by centrifugation at 10,000×g for 30 min and the clear supernatant was collected. It was further concentrated (50%) by Rotavapor rotary evaporation at 40°C under vacuum and subjected to hydrophobic column chromatography using Phenyl-Sepharose CL-4B (30 ml bed volume), previously equilibrated with 30% ammonium sulphate in 20 mM acetate buffer, pH 2.5. The column was washed thoroughly with 20 bed volumes of the above buffer and eluted with a 120 ml linear decreasing gradient of ammonium sulphate (30–0%) with a flow rate of 20 ml per hour and approximately 3.0 ml fractions were collected. Fractions showing activity at pH 2.5 (Phy I) and pH 5.0 (Phy II) were pooled separately, concentrated by rota vapor and loaded on a Sephacryl S-300 gel filtration column with a flow rate of 12 ml h−1 and 2 ml fractions were collected. Unless otherwise mentioned, all the purification procedures were carried out at 4°C.
Determination of molecular mass (Mr)
The molecular mass of purified enzyme was estimated by gel filtration, according to Andrews (1964). The gel filtration column of Sephacryl S-300 (1 × 150 cm) column equilibrated with 50 mM glycine-HCl buffer, pH 2.5 was calibrated using gel filtration standard molecular mass markers; cytochrome c (12.4 kDa), bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), apoferritin (443 kDa and thyroglobulin (669 kDa).
Electrophoretic techniques, mass spectrometry and N-terminal amino acid sequences analysis
Native PAGE (8%) was performed in a vertical gel apparatus at pH 7.5. Samples containing approximately 5–10 μg of protein were applied to the gel and electrophoresis was carried out at 200 V for 3–4 h. Protein bands were visualized by silver staining (0.4% w/v)(Deutscher 1990).
The subunit molecular mass was determined by SDS–PAGE in a vertical slab gel apparatus at pH 8.3 (Laemmli 1970) using Sigma high molecular mass markers. Isoelectric Focusing Polyacrylamide Gel Electrophoresis (IEF–PAGE) was performed in a tube gel (7.5%) using wide range Ampholytes (pH, 3–10) using a mini scale density gradient isoelectric focusing unit (Chinnatambi et al. 1994). Protein bands were detected either by Coomassie Brilliant Blue R-250 (0.2% w/v) or by silver staining (0.2% w/v) and phytase band was revealed by sodium phytate as a substrate.
Mass spectral analysis was performed on a Voyager-De-STR (Applied Biosystems) MALDI-TOF. A nitrogen laser (337 nm) was used for desorption and ionization. Spectra were acquired in the range of 10–100 kDa, in linear mode with delayed ion extraction and with an accelerating voltage of 25 kV. The low mass ion gate was set at 4,500 Da. All the analyses were performed in four replications. The instrument was calibrated with myoglobulin and bovine serum albumin. In-gel tryptic digestion and analysis of peptides was done according to Bar et al (2007).
For N-terminal sequence analysis, purified protein from the SDS–PAGE was electrotransfered to polyvinylidene difluoride membrane (PVDF, Bio-Rad, Richmond, CA, USA) using a semi-dry blotting system (Nova Blot, Pharmacia Biotech, Sweden) under a constant current of 52 mA for 30 min and the protein band was visualized by Coomassie Blue R-250 stain. The N-terminal amino acid sequence of the first 10 residues of the enzyme was determined by Edman degradation on an Applied Biosystems Procise sequencer at Commonwealth Biotechnologies, Inc., Richmond, Virginia, USA. Reported sequences of phytase from various sources were obtained from the GenBank and SwissProt Databases and a sequence alignment were performed using a Clustal-W sequence alignment program (Clustal W).
pH and temperature effect
The influence of pH on enzyme activity was determined by assaying phytase activity over the pH range 1.5–9.0, using 50 mM glycine-HCl buffer (pH, 1.5–4.5), acetate buffer (pH, 4.0–6.5) and Tris–HCl buffer (pH, 6.5–9.0) by the standard assay method. The pH stability was also determined in a same pH range by pre-incubating enzyme samples in above buffers at room temperature for a period of 12 h, aliquots were removed and immediately assayed for residual enzyme activity by the standard assay method.
The optimum temperature of phytase was determined by performing assays at temperatures 30–70°C. The temperature stability was also determined by incubating enzyme samples over a same temperature range for the period of 60 min. Aliquots (100 μl) were withdrawn after suitable time interval and residual enzyme activity was determined by the standard assay method.
Effect of various metal ions, sugars and inhibitor reagents
A suitably diluted enzyme (100 μl) in 50 mM Tris–HCl buffer, pH 7.0, was pre-incubated with various metal salts, sugars and inhibitor reagents at room temperature for 10 min and subsequently residual activity of the samples was determined by standard assay method.
Substrate specificity and kinetic studies
The relative substrate specificity of phytase towards various substrates was determined by varying the substrate concentrations from 0.1 to 10 mM. Assays were carried out at 55°C under standard assay conditions. The kinetic rate constants, K m , V max, K cat, and K cat /K m were determined graphically from Lineweaver and Burk plots (1934).
Amino acid analysis
The amino acid composition of purified Phy I and II was determined on a liquid phase automated amino acid analyser (Hewlett Packard Ti series 1050, equipped with a fluorescence detector) after hydrolysis in 6 M HCl for 20 h at 110°C. Total cysteine content was determined by the method of Habeeb (1972) and total tryptophan was determined according to Spande and Witkop (1967).
Results and discussion
Purification of phytases
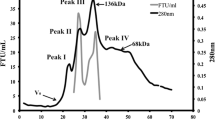
The purification of Phy I and Phy II was carried out as summarized in Tables 1 and 2. The crude culture supernatant was first concentrated (50%) on a Rotavapor and purified by adsorption and elution ion-exchange chromatography on Phenyl-Sepharose CL-4B, followed by gel filtration on Sephacryl S-300. Major activities were eluted in two peaks with a reverse gradient of ammonium sulphate (30–0%) in 20 mM acetate buffer, pH 2.5. The first peak was obtained at 15% ammonium sulphate with higher enzyme activity at pH 5.0 (Phy II), while the second peak was obtained at 5% ammonium sulphate concentration in 20 mM acetate buffer with higher enzyme activity at pH 2.5 (Phy I). These fractions were dialysed, concentrated and applied to a gel filtration column. The purified Phy I and II had a specific activity of 3436.4 and 1215.3 Umg−1 with 16.7- and 14.3-fold purification, respectively. It is reported in the literature (Ullah and Cummins 1987, 1988; Ullah and Gibson 1987) that A. niger NRRL 3135 phytases, viz., phyA, phyB, and pH 6.0 optimum acid phosphatase were secreted in relatively higher amounts under phosphate starvation conditions in starch medium, and only 5- to 25-fold purification was required to achieve near homogeneity using ion-exchange chromatography and chromatofocussing.
Determination of molecular mass, isoelectric point and glycosylation
The subunit molecular masses of purified Phy I and II were determined to be 66 and 150–160 kDa, respectively, by SDS–PAGE analysis (Fig. 1a), while that of native enzyme was found to be 264 and 150–160 kDa, respectively, by gel filtration on Sephacryl S-300 (Table 3). This suggests that Phy I is a tetramer and Phy II is a monomer. Dimeric Phy B has been reported from Aspergillus niger NRRL 3135 which has a molecular mass of 68 kDa (Ullah and Sethumadhavan 1988). The purity of phytases was further confirmed by IEF which showed a single band with pI value of ~3.65 and 3.95 for Phy I and Phy II, respectively (Fig. 1b). However MALDI data revealed the molecular weight of Phy I and II to be 66 and 74 kDa (Fig. 2). Thus the molecular weight of Phy II by MALDI-TOF was half the molecular weight shown by SDS gel electrophoresis. This could be because of the doubly charged species of Phy II during ionization process of MALDI-TOF spectrometry (Korwar et al. 2008).
a SDS–PAGE of purified Phy I and Phy II from Aspergillus niger NCIM 563. Lane M: Molecular weight markers. Lane 1: Crude filtrate (after rota vapour conc.). Lane 2: Phenyl Sepharose elute (with 5% w/v Ammonium sulphate). Lane 3: Phenyl Sepharose elute (with 15% w/v Ammonium sulphate). Lane 4: Purified Phy I. Lane 5: Purified Phy II. b Iso-electric focusing of purified Phy I and Phy II. Lane M: Standard pI markers (Pharmacia). Lane 1: Purified Phy I. Lane 2: Purified Phy II
MALDI-TOF analysis of peptides after in gel tryptic digestion of Phy I and Phy II. a Peptides of Phy I [Molecular weight of peptides (Da) 1563.3011, 2273.9826, 2304.9520, 2611.8672, 2928.8716, 3226.8563]. Inset: Molecular weight of Phy I by MALDI-TOF. b Peptides of Phy II [Molecular weight of peptides (Da) 2176.9194, 2214.9406, 2464.9820, 2693.8450, 3494.7825] Inset: Molecular weight of Phy II by MALDI-TOF
Effect of pH and temperature on phytases
Table 3 indicates that the optimum temperature of Phy I and II was 55°C but optimum pH was 2.5 and 5.0, respectively (Data not shown). Purified phytase from Aspergillus niger van Teighem also exhibited maximum activity at pH 2.5 and 52–55°C (Vats and Banerjee 2004). However phytase produced from Aspergillus niger CFR335 either under solid state or submerged fermentation showed phytase activity at pH 4.5 only (Gunashree and Venkateswaran 2008). Similarly Phy I had a half life of 6–8 h at 55°C while that of Phy II was 10–12 h. Several other microbial phytases (Konietzny and Greiner 2002) and various commercial phytases display an optimum temperature range of 45–55°C (Boyce and Walsh 2006). Phy I showed broad pH stability (pH 2–9) while Phy II was stable between pH 4.0 to 8.5 (Data not shown). After 65°C for 1.5 h, Phy I retained 40% activity while Phy II retained 55% of its original activity. However crude phytase from Aspergillus niger NCIM 563 was found to retain 100% of the original enzyme activity in the presence of 12% skim milk after treatment at 70°C for 3 min (Shah et al. 2009).
Effect of metal ions and other reagents on phytase activity
The effect of various metal ions (Fig. 3) and other reagents (Table 4) on Phy I and II were investigated. It was observed that Phy II activity was completely inhibited by Hg2+ at 1 mM concentrations, while Phy I retained 71% of its original activity. Fe3+ showed a negative effect (85%inhibition) on Phy I activity at 1 mM concentrations. Phy II was strongly inhibited by Cu2+ (retained 11%). The phytase from Saccharomyces cerevisiae was totally inhibited by Hg2+ and strongly inhibited by Fe2+ at 5 mM concentrations (In et al. 2009). The presence of EDTA stimulated Phy I activity, while showing slight inhibition of Phy II activity. Phytase from the marine yeast Kodamaea ohmeri BG3 also showed slight activation in phytase activity at concentrations up to 5 mM (Xiaoyu et al. 2008). However SDS showed negative effect on both Phy I and II. Oxalic acid and pCMB also showed a stimulating effect on Phy I activity (Table 4). Phytase from Aspergillus niger van Teighem, however, was not affected by most metal ions, inhibitors and organic solvents (Vats and Banerjee 2004).
Substrate specificity
The hydrolysis of several phosphate esters (3 mM) by purified Phy I and II was examined by measuring the release of free phosphate. The relative rate of hydrolysis of various substrates is summarized in Table 5. Phy I showed considerably broader substrate specificity and was noticeably active with other phosphate esters including p-nitrophenyl phosphate, a general substrate for acidic phosphatase. Similar results were also recorded for the phytase from Saccharomyces cerevisiae CY strain (In et al. 2009) and Mucor hiemalis (Boyce and Walsh 2007). Phytases derived from Aspergillus niger, namely Allzyme™ and Natuphos® also displayed broad substrate specificity (Martin et al. 2006). Many other fungal phytases also exhibit broad substrate specificity, which is desirable in terms of animal feed application (Wyss et al. 1999).
Kinetics
Results of the kinetic studies (Table 6) indicated that K m values for sodium phytate (2.01 and 0.145 mM, respectively) were lower than calcium phytate (37.27 mM & 1.14 mM, respectively) indicating that Phy I and II showed more affinity towards sodium phytate. However Phy I and ll showed higher V max (26,674 and 2,205 μmol min−1 mg−1 respectively) for calcium phosphate. Lower K m values (0.04–0.44 mM) for purified phytases from Aspergillus niger 92 and Aspergillus niger NRRL 3135 (Ullah and Gibson 1987) have been reported. Similarly among all substrates for phytase activity the turnover number (K cat ) values of both phytases (Phy I and II) was higher with calcium phytate as substrate. Furthermore the ratio of K cat /K m i.e. catalytic efficiency, showed that Phy II degraded sodium phytate and calcium phytate, 2.62- and 1.53-fold more efficiently than Phy I. The estimated K cat of the phytase I and II as computed from the kinetic linearity experiment was 2.2 × 104 and 4.2 × 103 s−1 with a catalytic efficiency of 1.1 × 104 and 2.8 × 104 M−1s−1 respectively. Despite the fact that the phytase I studied here showed a pH optimum similar to that of phyB, it showed marked differences in catalytic constants from the previously reported phytase (Ullah and Sethumadhavan 1998), which was found to possess a comparatively high affinity for phytate, with a K m of 0.103 mM and catalytic turnover of 628 s−1. However, the phytase studied here, although displaying a higher K m of 0.145 mM, was much more efficient in hydrolysing phytate, with a higher catalytic turnover and approximately 80% greater catalytic efficiency (6.1 × 106 M−1s−1) than phyB (Ullah and Sethumadhavan 1988). Phytase from Aspergillus niger van Teighem reported by Vats and Banerjee (2005) showed V max and K m 1,074 IU/ml and .606 mM, respectively, with a K cat 3 × 105 s−1 and catalytic efficiency of 3.69 × 108 M−1s−1.
N-terminal sequence
The N-terminal amino acid sequences of the first 10 amino acid residues of purified Phy I and II were found to be FSYGAAIPQQ and GVDERFPYTG, respectively. The comparison of the N-terminal amino acid sequence of Phy I and Phy II from other Aspergilli with other selected examples available in the literature is given in Table 7. The sequence of Phy II was different from known phytases. However, Phy I showed strong homology with the N-terminal sequence of Aspergillus ficuum pH 2.5 optimum acid phosphatase, which was later referred to as phyB phytase (Ehrlich et al. 1993; Vats and Banerjee 2006).
Amino acid analysis
The amino acid composition(Table 8) of the purified phytases I and II revealed the enzymes to have 2,471, 1,448 amino acid residues and a calculated molecular weight of about 271.81 and 159.29 kDa respectively. The content of hydrophobic or apolar amino acids (leucine, isoleucine, valine, methionine, proline, alanine, phenylalanine, tryptophan) of Phy I and II is about 40.3 and 38.4% (residues/mol). Polar amino acids (Gly, Ser, Thr, Cys, Asx, Glx, Tyr) content is 52.6 and 53.3%. The Asx and Glx content is about 22.5 and 19.8% and outnumbered the basic amino acids (Lys, Arg, His) about 7.1 and 8.3%. The enzymes had higher proline content (6.0 and 6.2%). The deduced amino-acid sequence is currently available for three phyB genes: phyB from A. niger NRRL 3135 (Ehrlich et al. 1993), aph from A. niger var. Awamori ATCC 38854 (Piddington et al. 1993), and pH-2.5 acid phosphatase A. niger T213 (Kostrewa et al. 1999).
Conclusion
Earlier Fujita et al. (2003) reported two types of phytase from Aspergillus oryzae under solid state fermentation conditions which showed the same pH optima (5.0), thermostability (100% stable up to 50°C) but differed in their temperature optima (65 and 55°C, respectively) and molecular weight (120 and 116 kDa, respectively). Extracellular phytase with two pH optima (2.5 and 5.0) has been reported from Aspergillus niger (Ullah 1988) and commercial Aspergillus niger phytase (Natuphos) (Wyss el al. 1999). We have earlier reported production and partial characterization of two types of phytase from Aspergillus niger NCIM 563 under submerged fermentation (Soni and Khire 2007). In the present communication we were able to purify both phytases (Phy I active at pH 2.5 and Phy II active at pH 5.0) from Aspergillus niger NCIM 563 produced under submerged fermentation to homogeneity and found that they differed from each other in molecular and biochemical properties including pH optima, temperature and pH stability, effect of various metal ions and other reagents, MALDI-TOF analysis, affinity towards various substrates and N-terminal amino acid analysis. This, according to our knowledge, is the first report of two distinct phytases from Aspergillus niger produced under submerged fermentation conditions.
The novelty of Phy I with highly acidic pH optimum (2.5), stability under wide pH range (1.5 to 9.0) and broad substrate specificity suggests its application in poultry feed. However, the further work needed to improve its thermostability by immobilization and/or micro encapsulation along with site directed mutagenesis for its application in pelleted poultry feed is in progress.
References
Andrews P (1964) Estimation of the molecular weights of proteins by Sephadex gel filtration. Biochem J 91:222–233
Bar C, Patil R, Doshi J, Kulkarni MJ (2007) Characterization of the proteins of bacterial strains isolated from contaminated site involved in heavy metal resistance-A proteomic approach. J Biotechnol 128:44–451
Bhavsar K, Shah P, Soni SK, Khire JM (2008) Influence of pretreatment of agriculture residues on phytase production by Aspergillus niger NCIM 563 under submerged fermentation conditions. African J Biotechnol 7:1101–1106
Boyce A, Walsh G (2006) Comparision of selected physicochemical characteristics of commercial phytases relevant to their application in phosphate pollution abatement. J Enviorn Sci Health Part A: Toxic/Hazard Subst Enviorn Eng 41:789–798
Boyce A, Walsh G (2007) Purification and characterization of an acid phosphatase with phytase activity from Mucor hiemalis. J Biotechnol 132:82–87
Chinnatambi S, Bodhe AM, Lachke AH (1994) Fabrication of improved mini scale eletrofocusing unit. Biotechnol Tech 8:681–682
Deutscher MP (1990) Guide to protein purification. Methods Enzymol 182:529–539
Ehrlich KC, Montalbano BG, Mullaney EJ, Dischinger HC, Ullah AHJ (1993) Identification and cloning of a second phytase gene (phyB) from Aspergillus niger (ficuum). Biochem Biophys Res Commun 192:747–753
Fujita J, Yamane Y, Fukuda H, Kizaki Y, Wakabayashi S, Shigeta S, Suzuki O, Ono K (2003) Production and properties of phytase and acid phosphatase from a sake koji mold, Aspergillus oryzae. J Biosci Bioeng 95:348–353
Gunashree BS, Venkateswaran G (2008) Effect of different cultural conditions for production of phytase by Aspergillus niger CFR 335 under submerged and solid state fermentations. J Ind Microbiol Biotechnol 35:1587–1596
Habeeb AFSA (1972) Reaction of protein sulfhydryl groups with Ellman’s reagent. Methods Enzymol 25:457–464
Heinohen JK, Lathi RJ (1981) A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem 113:313–317
In MJ, Seo SW, Kim DC, Oh NS (2009) Purification and biochemical properties of an extracellular acid phytase produced by the Sachharomyces cerevisiae CY strain. Process Biochem 44:122–126
Kaur P, Kunze G, Satyanarayana T (2007) Yeast phytases: present scenario and future perspectives. Critical Rev Biotechnol 27:93–109
Kim YO, Kim HK, Baeks YJ, Oh TK (1998) Purification and properties of a thermostable phytase from Bacillus spp DS11. Enzyme Microb Technol 22:2–7
Konietzny U, Greiner R (2002) Molecular and catalytic properties of phytase- degrading enzymes (phytases). Int J Food Sci Technol 37:791–812
Korwar AM, Santhakumari B, Kulkarni MJ (2008) Identification of ripening specific proteins in tomato by intact tissue MALDI-TOF-MS. Electronic J Food Plants Chem 3:10–13
Kostrewa D, Wyss M, D’Arcy A, van Loon APGM (1999) Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2.4 A resolution. J Mol Biol 288:965–974
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680–685
Lineweaver H, Burk D (1934) Determination of enzyme dissociation constants. J Amer Chem Soc 56:656–658
Martin JA, Murphy RA, Power RFG (2006) Purification and physico- chemical characterization of genetically modified phytases expressed in Aspergillus awamori. Bioresour Technol 97:1703–1708
Mullaney EJ, Daly C, Ullah AB (2000) Advances in phytase research. Adv Appl Microbiol 47:157–199
Piddington CS, Houston CS, Paloheimo M, Cantrell M, Meittinen-Oinonen A, Nevalainen H, Rambosek J (1993) The cloning and sequencing of the genes encoding phytase (phy) and pH 2.5-optimum acid phosphatase (aph) from Aspergillus niger var. awamori. Gene 133:55–62
Radcliffe JS, Zhang Z, Kornegay ET (1998) The effect of microbial phytase, citric acid and their interaction in a corn soybean meal based diet for Weaning pigs. J Animal Sci 76:1880–1886
Selle PH, Ravindran V (2007) Microbial phytase in poultry nutrition. Animal Feed Sci Technol 135:1–41
Shah P, Bhavsar K, Soni SK, Khire JM (2009) Strain improvement and up scaling of phytase production by Aspergillus niger NCIM 563 under submerged fermentation conditions. J Ind Microbiol Biotechnol 36:373–380
Singh PK (2008) Significance of phytic acid and supplemental phytase in chicken nutrition: a review. World’s Poultry Sci J 64:553–580
Soni SK, Khire JM (2007) Production and partial characterization of two types of phytase from Aspergillus niger NCIM 563 under submerged fermentation conditions. World J Microbiol Biotechnol 23:1585–1593
Spande TF, Witkop B (1967) Determination of Tryptophan content of protein with N-bromosuccinimide. Methods Enzymol 11:498–506
Ullah AHJ (1988) Production, rapid purification and catalytical characterization of extracellular Phytase from Aspergillus ficuum. Prep Biochem 18:443–458
Ullah AHJ, Cummins BJ (1987) Purification, N-terminal amino acid sequence and characterisation of pH 2.5 optimum acid phosphatase (E.C.3.1.3.2) from Aspergillus ficuum. Prep Biochem 17:397–422
Ullah AHJ, Cummins BJ (1988) Aspergillus ficuum extracellular pH 6.0 optimum acid phosphatase: purification, N-terminal amino acid sequence and biochemical characterization. Prep Biochem 18:37–65
Ullah AHJ, Gibson DM (1987) Extracellular phytase (EC 3.1.3.8) from Aspergillus ficuum NRRL 3135: purification and characterization. Prep Biochem 17:63–91
Ullah AHJ, Sethumadhavan K (1988) Differences in the active site environment of Aspergillus ficuum phytases. Biochem Biophys Res Commun 243:458–462
Vats P, Banerjee UC (2004) Production studies and catalytic properties of phytases (myoinositolhexakisphosphate phosphohydrolases): an overview. Enzyme Microb Technol 35:3–14
Vats P, Banerjee UC (2005) Biochemical characterisation of extracellular phytase (myo-inositol hexakisphosphate phosphohydrolase) from a hyper- producing strain of Aspergillus niger van Teighem. J Ind Microbiol Biotechnol 32:141–147
Vats P, Banerjee UC (2006) Catalytic characterization of phytase (myo- inositolhexakisphosphate phosphohydrolase) from Aspergillus niger van Teighem: Glycosylation pattern, kinetics and molecular properties. Enzyme Microbial Technology 39:596–600
Vats P, Sahoo DK, Banerjee UC (2004) Production of phytase (myo- nositolhexakis phosphate phosphohydrolase) by Aspergillus niger van Teighem in laboratory scale fermenter. Biotechnol Prog 20:737–743
Vohra A, Satyanarayana T (2003) Phytases: microbial sources, production, purification and potential biotechnological applications. Critical Rev Biotechnol 23:29–60
Wyss M, Brugger R, Kronenberger A, Remy R, Oesterhelt G, Lehman M, van Loon APGM (1999) Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolase): catalytic properties. Appl Enviorn Microbiol 65:367–373
Xiaoyu L, Zhenming C, Zhiqiang L, Jing L, Xianghong W, Hirimuthugoda NY (2008) Purification and characterization of extracellular phytase from a Marine yeast Kodamaea ohmeri BG3. Marine Biotechnol 10:190–197
Clustal W Multiple sequence alignment program. http://www.ebi.ac.uk/clustalw/
Acknowledgments
The authors gratefully acknowledge financial support provided by Department of Science and Technology, New Delhi, India. One of the authors, Mr. S. K. Soni thanks Council of Scientific and Industrial Research (CSIR), India for financial assistance. We also acknowledge the assistance from Dr. Mahesh Kulkarni for MALDI-TOF analysis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Soni, S.K., Magdum, A. & Khire, J.M. Purification and characterization of two distinct acidic phytases with broad pH stability from Aspergillus niger NCIM 563. World J Microbiol Biotechnol 26, 2009–2018 (2010). https://doi.org/10.1007/s11274-010-0385-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-010-0385-8