Abstract
Chaihu, the dried roots of several Bupleurum species, are widely used as herbal drugs. Triterpene saponins, as saikosaponins (SSs) in Chaihu, exhibit a wide range of pharmacological actions, including significant anti-inflammatory, immunomodulatory, hepatoprotective, antioxidant, anti-tumor and antiviral activities. To explore the regulatory biosynthesis of SS in Bupleurum chinense roots, we screened putative transcription factor (TF) genes from transcriptome data. Here, we report one of the bZIP TF genes, named BcbZIP134, which belongs to bZIP G subfamily. The expression of BcbZIP134 was elevated in MeJA-treated adventitious roots of B. chinense. No obvious differential expression of BcbZIP134 was detected in tissues (root, stem, leaf, flower and fruit) of B. chinense. Overexpressing BcbZIP134 in B. chinense hairy roots revealed that the content of the main SS monomers a, c and d were decreased compared to control hairy roots. Furthermore, the contents of a few other minor SS monomers were also affected when compared to the controls. The results indicate that BcbZIP134 may play a negative regulatory role in SS biosynthesis.
Key Message
Chaihu, the dried roots of several Bupleurum species, are successfully and widely used as herbal drugs. Triterpene saponins, as saikosaponins (SSs) in Chaihu, exhibit a wide range of pharmacological actions, including significant anti-inflammatory, immunomodulatory, hepatoprotective, antioxidant, anti-tumor and antiviral activities. To explore the regulatory biosynthesis of SS in B. chinense roots, we screened putative transcription factor (TF) genes from transcriptome data. Here, we report one of the bZIP TF genes, named BcbZIP134, which encodes a predicted 410 amino acid protein belonging to bZIP G subfamily. The expression of BcbZIP134 was elevated in MeJA-treated adventitious roots of B. chinense, and no obvious differential expression of BcbZIP134 was detected in tissues (root, stem, leaf, flower and fruit) of B. chinense. Overexpressing BcbZIP134 in B. chinense hairy roots revealed that the content of the main SS monomers a, c and d were decreased compared to control hairy roots. Furthermore, the contents of a few other minor SS monomers were also affected when compared to the controls. The results indicate that BcbZIP134 may play a negative regulatory role in SS biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
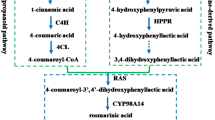
Saikosaponins (SSs), which are triterpene saponins, are the main active components in Radix Bupleuri, the roots of some species from the genus Bupleurum. To date, more than 120 glycosylated oleanane-type and ursane-type saponins have been isolated from Bupleurum. The predominant SS monomers in Bupleurum are SS a, SS c and SS d, of which structures are shown in Fig. 1. Isolated SS monomers such as SS a, SS c, SS d, SS b1 and SS b2 or an SS mixture displayed pharmacological activity on Hepatitis B replication, acute and chronic inflammation, T cells and B cells proliferation, lipid peroxidation and PGE2 production (Ashour et al. 2011). The putative SS biosynthetic pathway initiates the isoprenoid pathway, mediates the cyclization of oxidosqualene and then undergoes some modifications of oxidation, glycosylation and other secondary transformations (Lin et al. 2013). The genes encoding some of the biosynthetic enzymes were cloned mainly from B. chinense, B. falcatum and B. kaoi (Gao et al. 2015; Moses et al. 2014; Chen et al. 2007; Kim et al. 2006). Previous studies have shown that some endogenous signal molecules, such as methyl jasmonate (MeJA) and abscisic acid (ABA), or some exogenous fungal elicitors could elevate the biosynthesis of SSs (Chen et al. 2007; Aoyagi et al. 2001; Kim et al. 2011). When potted B. chinense seedlings were subjected to progressive drought stress, both SS a and SS d content increased and were accompanied by an enhanced content of active oxygen species (Zhu et al. 2009a, b). However, the molecular regulatory mechanism of the SS biosynthesis remains elusive.
Regarding the regulation of plant terpenoid biosynthesis, a few transcription factors (TFs) belonging to different families have been identified. For example, AabZIP1, AaWRKY1, AaERF1 and AaERF2 from Artemisia annua were shown to regulate the biosynthesis of the sesquiterpenoid artemisinin. AabZIP1 activated the expression of the two key genes encoding amorpha-4,11-diene synthase (ADS) and CYP71AV1 and its activation was enhanced by ABA treatment (Zhang et al. 2015a, b). AaWRKY1 bound to the W-box of the promoter of the ADS gene and activated its expression (Ma et al. 2009). AaERF1 and AaERF2 bound to the CRTDREHVCBF2 and RAV1AAT motifs present in both ADS and CYP71AV1 promoters, and the overexpression of either of these TFs resulted in increased accumulation of artemisinin and artemisinic acid (Yu et al. 2012). CrMYC2, ORCA2, CrGBF1 and CrGBF2 were TFs which were found regulating the biosynthesis of terpenoid indole alkaloids in Catharanthus roseus (Siberil et al. 2001; Memelink et al. 2007; Zhang et al. 2011a, b) In addition, several TFs regulating the triterpene (saponin) biosynthesis have been identified. In Medicago truncatula, two homologous jasmonate-inducible TFs of the basic helix-loop-helix family (bHLH), Triterpene saponin biosynthesis activating regulator1 (TSAR1) and TSAR2, were found directly binding to the N-box in the promoter of the gene encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 (HMGR1) to enhance triterpene saponin biosynthesis (Mertens et al. 2016). Another bHLH TF in glycyrrhiza uralensis, GubHLH3, was shown to regulate the expression of soyasaponin biosynthetic genes positively (Tamura et al. 2018).
The basic leucine zipper (bZIP) is a large family of TFs in plants and the members of this family contain a conserved DNA-binding basic region and a dimerized leucine zipper region. Diverse roles have been found for bZIP TFs especially in plant stress response and hormone signal transduction (Hurst et al. 1995; Llorca et al. 2014). The bZIP-family in Arabidopsis thaliana was sorted into 13 groups comprising 78 members. The members in the same groups tended to have similar structural features and functions (Dröge-Laser et al. 2018). In addition to terpenoids, bZIP TF members were identified by their regulation of the biosynthesis of other metabolites, such as HY5 and HYH from A. thaliana, regulating the synthesis of anthocyanins and improving the cold resistance by regulating the key gene DFR of the anthocyanin synthesis pathway (Zhang et al. 2011a, b).
Bupleurum chinense DC. is the dominant source species of Radix Bupleuri and has been domesticated for several decades in China. To explore the biosynthesis and regulation of SSs in B. chinense, we previously analyzed the transcriptome of B. chinense intending to search for genes that may participate in the biosynthesis of SSs (Sui et al. 2011). The transcriptomes of the roots of B. chinense and another Bupleurum species, B. scorzonerifolium, were compared to find clues about the regulation of SS biosynthesis because SSs are easily detected in roots of the former while hardly detected at all in roots of the latter (Sui et al. 2014). A number of genes encoding TFs were found to be exceptionally expressed in only one of the two species or were distinctly expressed in both species. In this study, the impact on SS biosynthesis of one of the TFs genes, BcbZIP134, was further explored. BcbZIP134 was overexpressed in the hairy roots of B. chinense by Agrobacterium rhizogenes-intermediated transformation. The SS compounds were assayed by high performance liquid chromatography quadrupole/time of flight-mass spectrometry/mass spectrometry (HPLC–QTOF–MS/MS).
Materials and methods
Candidate TF genes selection
To identify candidate TF genes that may be involved with SS biosynthesis (The putative biosynthetic pathway presented in our previous report (Sui et al. 2011)), we first analyzed the category of the TFs based on their sequences obtained from the transcriptome data of B. chinense and B. scorzonerifolium. Those that differentially expressed in the roots of both species and that belonged to the families of AP2, bZIP and bHLH were selected for further screening. A total of 26 TF genes, including 18 AP2 genes, four bZIP genes and four bHLH genes, were analyzed by expression profiling in MeJA-treated B. chinense adventitious roots by quantitative real time polymerase chain reaction (qRT-PCR). Next, the contents of SS a, SS c and SS d were assayed by HPLC. Six TF genes, including BcbZIP134, exhibited elevated expression accompanied by enhanced levels of SSs. Then, the expression of the six genes in roots, stems, leaves, flowers and fruits of B. chinense were tested by qRT-PCR. The experimental protocols of adventitious root culture and qRT-PCR were done according to previous reports (Sui et al. 2014; Kusakari et al. 2000). In addition, the time course of the expression of these genes after MeJA treatment was assayed. MeJA was added to the media at a final concentration of 200 µM when adventitious roots were transferred to the second step culture media (Aoyagi et al. 2001). The roots were collected at 0 h, 2 h, 4 h, 8 h, 12 h, 24 h, 48 h, 72 h and 5 days after MeJA treatment and were immediately frozen in liquid nitrogen then stored at − 80 °C until needed. Roots in six bottles were treated as one replicate and three replicates were prepared. In addition, the collection of tissues from B. chinense was done according to our previous report (Dong et al. 2011).
Extraction of SSs and HPLC Analysis
The adventitious roots were dried for 72 h using a freeze-drier (LGJ-18, Beijing Songyuan Huaxing Technology Develop Co. Ltd., China). Powdered dried roots (0.5 g) of each sample were ultrasonically extracted with 25 mL of a 5% ammonia/methanol solution in 100 mL flasks for 30 min. The extract was filtered and then evaporated until dry using 70 °C water baths. The residue was dissolved in chromatographic methanol and adjusted to 5 mL using a volumetric flask. Before injection, the solution was filtered using a 0.45 µm microfiltration membrane. The reference standards of SS a, SS c and SS d were purchased from the National Institutes for Food and Drug Control, Beijing, China. The HPLC (Waters 1525 Binary HPLC Pump, USA) and an ASB-vensil C18 column (250 mm × 4.6 mm, 5 µm) were used. A gradient elution of A (water/0.05% NH4OH) and B (acetonitrile) was used as follows: 0–15 min, 10–85% B; 15–18 min, 85% B; 18–22 min, 85–10% B; and 22–27 min, 10% B. The detection was set at 210 nm. The injection volume was 20 µL, the flow rate was 1.5 mL/min and the column temperature was 40 ± 0.15 °C. The total content of SS a, SS c, and SS d was calculated for comparison because these three SSs are the dominant SSs in B. chinense.
Full-length cDNA cloning and bioinformatic analysis of BcbZIP134
To verify the sequence of BcbZIP134 from transcriptome data and to obtain the full-length cDNA sequence, rapid amplification of cDNA ends (RACE) PCR was used to amplify the 5ʹ and 3ʹ ends of the sequences using a SMARTer™ RACE cDNA Amplification kit (Clontech Laboratories, CA, USA). Then, the full-length cDNA was amplified using specific primers in one PCR reaction. PCR amplification conditions for the 3′ and 5′ RACE were as follows: five cycles at 94 °C for 30 s and 72 °C for 3 min; five cycles at 94 °C for 30 s, 70 °C for 30 s, and 72 °C for 3 min; 25 cycles at 94 °C for 30 s, 68 °C for 30 s, and 72 °C for 3 min; a final extension for 10 min at 72 °C; and then cooled to 4 °C. All primers were designed using Primer Premier 5.0 (Primer Biosoft International, Palo Alto, California, USA) and are listed in the Appendix. Search for similar sequences of BcbZIP134 was carried out using the blastx suite at the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The amino acid sequences of hits with high scores were downloaded and then were used to construct a phylogenetic tree using the MEGA7 software. Alignment of these sequences was conducted using DNAman in order to show the conserved motifs and identity between BcbZIP134 and another single sequence.
Agrobacterium rhizogenes-intermediated transformation of BcbZIP134
Bupleurum chinense seeds were grown on MS solid medium, after surface sterilization in 20% sodium hypochlorite solution for 15 min. The tender leaves of sterile seedlings were used for A. rhizogenes infection. A single colony of A. rhizogenes ACC10060 strain carrying the pK2GW7 plasmid with the BcbZIP134 and a spectinomycin resistance gene (NptII) was inoculated into 5 mL of lysogeny broth (LB) medium supplemented with spectinomycin (50 mg/L) and rifampin (50 mg/L) and cultured overnight at 28 °C with shaking (180 rpm). Then, the cultures were expanded into a larger volume, and culturing was continued until the OD600 was approximately 0.8. The cultures were centrifuged for 10 min at 5000 rpm at 4 °C and were resuspended in equal volume of MS liquid medium containing 200 µM acetosyringone. The explants were dipped into the infection solution for 20 min and were cocultured on Murashige and Skoog (MS) medium at 25 °C in the dark for 3 days after being blotted dry on sterile filter paper. The explants were then transferred to MS medium containing 500 mg/L of cefotaxime. The hairy roots started to develop after about 2 weeks of cultivation. Hairy roots of approximately 2 cm in length were excised from explants and cultured in fresh MS medium containing 500 mg/L cefotaxime in the dark. After approximately 4 weeks, these hairy roots were transferred to MS liquid medium, cultured in the dark with shaking (150 rpm) and maintained as separate independent lines. The roots were transferred to fresh medium once every 4 weeks until the quantity of each independent line was enough for HPLC analysis. The hairy root lines that were induced by nontransformed A. rhizogenes were used as controls. The transgenic hairy root lines were verified by PCR amplification with primers for 35 S promoter and 35 S terminator in the vector. The overexpression of BcbZIP134 was evaluated by real-time qRT-PCR.
SSs profile analysis in BcbZIP134-overexpressed hairy root lines
Profiles of the SSs were analyzed for five independent lines overexpressing BcbZIP134. The SS extraction method was identical to the method used to extract SSs from adventitious roots of B. chinense described above, except a procedure of solid-phase microextraction (SPME) was added. An SPME column (Bonna-Agela Cleanert S C18, China) was used. First, the column was activated with 5 mL of methanol followed by 5 mL of chromatographic water washing. The materials in 20% methanol were slowly added to the activated SPME column, and, after the sample was completely loaded into the column, 3 mL 20% aqueous methanol solution was used for washing and then the column was vacuum dried. Then, an 85% aqueous methanol solution was used to elute with a final volume of 2 mL. Finally, the eluted solution was blow-dried with nitrogen and dissolved into 500 µL of methanol for assay by HPLC–QTOF–MS/MS (Agilent APCI–QTOF).
HPLC separation was performed on an Agilent Zorbax Extend-C18 (1.8 µm 2.1 × 100 mm) column using an Agilent 1260 Series system (Agilent, USA). The total flow rate was 0.2 mL/min, and the total elution program was 36 min with an optimized linear gradient elution of A (5 mM ammonium acetate) and B (acetonitrile) as follows: 0–15 min, 15–85% B; 15–20 min, 85% B; 20–25 min, 85–100% B; 25–31 min, 100% B; 31–31.1 min, 100–10% B; and 31.1–36 min, 10% B. The column temperature was maintained at 35 °C. The injection volume was 50 µL.
Mass spectra were acquired with an Agilent Accurate-Mass-Q-TOF MS 6520 system equipped with an atmospheric chemical ionization (APCI) source. All MS experiments were detected in the positive ionization mode. For Q-TOF/MS conditions, fragmentor and capillary voltages were kept at 240 and 3500 V, respectively. The temperature of the drying gas was set at 300 °C. The flow rate of the drying gas and the pressure of the nebulizer were 10 L/min and 25 psi, respectively. Full-scan spectra were acquired over a scan range of m/z 80–2500 at 1.03 spectra/s.
The peaks on the representative total ions current chromatograms representing SS a, SS c and SS d were identified by comparison of their chromatographic retention times, accurate molecular weights and characteristic mass fragment ions with those of the references standards. Other peaks were tentatively identified by comparison of their accurate mass data with those reported in the literature. Detailed information related to the illustration of peaks which showed variation between transformed and control samples is shown in Table 1. The quantification of SS a, SS c and SS d was calculated by comparison with reference standards. In addition, the relative quantification of other SS monomers was evaluated by comparison of the peak areas between those in transformed and control samples.
Results
Screening and cloning of BcbZIP134
A total of 26 TF genes were assayed for their expression profiles, and the results were combined with the SSs content assays in MeJA-treated B. chinense adventitious roots. Among those TF genes, the expression of BcbZIP134 was elevated after MeJA treatment (Fig. 2a). The significant elevation started at 24 h after MeJA treatment but gradually decreased beginning 72 h after treatment. This changing tendency was slightly ahead of the rapid increase in the accumulation SSs (Fig. 2b). The expression of BcbZIP134 in the roots, stems, leaves, flowers and fruits were tested and the results did not exhibit any remarkable differences among tissues (Fig. 2c). Except for BcbZIP134, expressions of five other TF genes were enhanced by MeJA treatment. We present here the relevant results of BcbZIP134. Further exploration in the function of the five TF genes are underway and will be reported later.
Expression of BcbZIP134 in MeJA-treated adventitious roots and different plant tissues. Expression of BcbZIP134 (a) and the content of SSs (b) at 0, 2, 4, 8, 12, 24, 48, 72 h and 5 days after 200 µM MeJA added into the medium. The relative expression was calculated using the 2−ΔΔCt formula with the expression in nontreated roots (0 h) as the calibrator. The content of SSs was calculated as the sum of SS a, SS c and SS d. c Expression of BcbZIP134 in roots, stems, leaves, flowers and fruits was calculated using the 2−ΔCt formula
Sequence character of BcbZIP134
The full-length cDNA of BcbZIP134 was generated using RACE PCR. It included an open reading frame of 1,230 bp, encoding a predicted 410 amino acid protein. Analysis of the deduced amino acid sequence revealed that this protein had one typical bZIP TF DNA binding domain at aa 288–351. The sequence alignment showed that BcbZIP134 had high sequence identity with bZIP TF 16-like and HBP-1a proteins, which belong to the G group of bZIP TFs from various plant species, such as Daucus carota subsp. Sativus, Petroselinum crispum, Citrus clementina, and Coffea canephora. Definite functions of these proteins homologous to BcbZIP134 are still unknown. The phylogenetic tree was shown as Fig. 3 and the alignment was shown as Fig. 4. Similar to other members of the G group of bZIP TF, BcbZIP134 had characteristic domains 1, 2, 3 which are parts of a proline-rich activation domain, as well as a bZIP domain (labeled in Fig. 4).
The phylogenetic tree of BcbZIP134 and its homologous proteins was constructed by MEGA7. Alignment of all sequences to show the conserved motifs was presented in Fig. 4
Alignment of the amino acid sequence of BcbZIP134 and its homologous proteins from other plants. The conserved motifs are labeled. Motifs one, two and three were identified according to a reference (Jakoby et al. 2002) as part of a proline-rich activation domain in bZIP group G. The identity of each protein with BcbZIP134 is provided in the brackets after the protein name. The plant source and GenBank accession numbers of all these sequences were shown in Fig. 3
Overexpression of BcbZIP134 in hairy roots of B. chinense
The plant overexpression vector pK2GW7 harboring BcbZIP134 driven by the 35S promoter was transferred into A. rhizogenes for hairy roots generation. The overexpressing hairy root lines were screened using real-time qRT-PCR and five independent lines, 19, 43, 54, 55 and 60, with considerably overexpressed BcbZIP134 transcript levels selected for the SSs contents assay (Fig. 5a). Compared with control hairy roots, BcbZIP134-overexpressing transgenic hairy roots grew slightly faster but stouter, and no difference in color appearance. The content of SS a, SS c and SS d, the main monomers of SS, were calculated and compared between BcbZIP134-overexpressed lines and controls. The content of SS a showed significant decrease in all of the overexpressed lines (Fig. 5c). The content of SS c in two lines 19 and 60, and the content of SS d in three lines 19, 55 and 60 significantly decreased (Fig. 5d, e). In all, the total content of SS a, SS c and SS d decreased significantly in all of the overexpressed lines (Fig. 5b).
Transcript abundance and SS content in B. chinense BcbZIP134-overexpressing hairy roots lines and controls. a Transcript abundance showing significantly elevated BcbZIP134 levels in the overexpressed lines. b Comparison of the total content of SS a, SS c and SS d showing significant decreases in the overexpressed lines. c–e The individual contents of SS a, SS c and SS d are compared showing altered changes in different overexpressed lines. Hairy root samples collected at three times (20 days, 25 days, and 30 days after changing new culture media) during growth were treated as three replicates and the symbol (*) represents statistical significance by Student’s t tests
In addition to the major SS a, SS c and SS d components, other minor SS monomers predicted by previous references were also assayed. One unknown compound (C37H62O7, compound 7 in Table 1) only existed in the controls. Five compounds (C48H78O18 unknown compound 1; C45H70O18 unknown compound 3; C44H70O14 acetyl-SS a compound 5; C42H68O12 SS e compound 6 and C44H70O14 acetyl-SS a compound 8 in Table 1) had significantly lower content in certain BcbZIP134-overexpressed lines than in the controls, and due to the large diversity seen between samples within the same line, the high standard deviation for these lines caused the statistical nonsignificance. The content of two compounds (C44H70O14 acetyl-SS a compound four and C44H70O14 acetyl-SS d compound 9) was decreased but was not statistically significant. Only lines 43 and 54 had an increased content of compound 2 (C44H70O14 acetyl-SS a or -SS d), which was the opposite of the others, showing inconsistent variation among individual lines. As a whole, the overexpression of BcbZIP134 caused a significant decrease of the total SS content in hairy roots, especially SS a, which may play a negative regulatory role in the SS synthesis pathways.
Discussion
In this study, hairy root transgenic systems were used to explore gene function. Unlike many other plants, most medicinal plants lack a transgenic model system, and this has hindered research progress on functional genes. Hairy root transgenic systems have been used as an effective model in a few medicinal plant species and other crops, such as ginseng (Zhang et al. 2015a, b), Danshen (Xu et al. 2017), Scutellaria baicalensis (Zhao et al. 2018), and M. truncatula (Mertenes et al. 2016). These systems are practical, considering the long growth period of the roots and since roots are used as the therapeutic parts for most medicinal plant species. In addition, a recent study reported the production of sapogenins (stigmasterol and hecogenin) using genetically transformed hairy roots of Chlorophytum borivilianum (Bathoju et al. 2017). Here, for the first time, we report the use of transgenic hairy roots of B. chinense to study gene functions. We believe this system will promote biological and genetic studies on this herb that is used in both traditional and modern medicine. When we overexpressed the cDNA of BcbZIP134 using the hairy root transgenic system, negative regulation function of this TF on SS biosynthesis was deduced. While in our primary screening with MeJA treated adventitious roots, both expression of BcbZIP134 and content of SSs were elevated. This suggests a positive regulation function of BcbZIP134 to the SS biosynthesis. It implied that the increase of SSs may be regulated by multiple ways and it is the combined results of different regulations. The relationship among SS biosynthesis and MeJA-responsive genes needs to be explored further.
The biosynthesis of secondary metabolites can be affected by developmental and environmental conditions. Although dozens of different SS monomer compounds have been detected in the roots of Bupleurum, little investigation has been conducted on the biosynthesis and regulation of different SS monomers. This is especially true for minor compounds and their mutual transformation in a given Bupleurum species (Li et al. 2015). In this study, we compared the content of SS a, SS c and SS d, the three major SS monomers in Bupleurum, with reference standard compounds. Other minor compounds were assayed based on the literature reports (Zhu et al. 2017; Fang et al. 2017). Nine minor compounds showed content variation (listed in Table 1). Other SS compounds such as SS f, SS h, SS i and SS b were not detected in either BcbZIP134-overexpressed lines or controls. This may indicate that differences exist between hairy roots and other plant roots, or that differences exist in hairy roots cultivated for different lengths of time. We have noticed that the hairy roots cultivated for 2 months after new culture media was changed usually synthesized more SS. Additionally, seven of the nine compounds were significantly decreased in transgenic line 19, combined with the lowest content of SS a, SS c and SS d in it, which showed the transgenic line 19 was most affected. While the transgenic line 54 showed most abundance of transcripts compared with other transgenic lines. It did not correspondingly possess the lowest content of SSs, although significant decrease in total contents of SS a, SS c and SS d were observed. It indicates that some unknown mechanism may exist to fine tune the accumulation of SSs in B. chinense and this mechanism functions when SS biosynthesis is severely threatened.
It has been reported that salt and drought stress can potentially increase pharmaceutically significant secondary compounds in plants (Selmar et al. 2008; Nasrollahi et al. 2014) and that drought stress, fertilizer or altered planting time influences the SS a, SS c and SS d content in Bupleurum roots (Zhang et al. 2016; Lee et al. 2002; Zhu et al. 2009a, b). The underlying molecular regulatory mechanisms of these changes have seldom been explored. As for other plant species, a few TFs have been reported to regulate the biosynthesis of terpenoids. Those belonging to the bZIP family include AabZIP1 from Artemisia annua (Zhang et al. 2015a, b) and OsTGAP1 and OsbZIP79 from Oryza sativa (Okada et al. 2009; Miyamoto et al. 2014). OsTGAP1 exerts an essential role on the expression of the clustered genes for momilactone biosynthesis and functions as a key regulator of the coordinated transcription of genes involved in inductive diterpenoid phytoalexin production in rice (Okada et al. 2009). OsbZIP79 interacts with OsTGAP1 and functions as a negative regulator of diterpenoid phytoalexin production that is triggered by a chitin oligosaccharide elicitor in rice cells (Miyamoto et al. 2014) It may be speculated TFs play vital linking roles between stress responses and secondary metabolism in plants and much related to be discovered.
Among the A. thaliana bZIP-family, Group G contains five members: AtbZIP16, AtbZIP68, AtbZIP41 (AtGBF1), AtbZIP54 (AtGBF2) and AtbZIP55 (AtGBF3) (Dröge-Laser et al. 2018). The amino acid sequence of BcbZIP134 shares a high level of identity with bZIP TF 16-like proteins. It has been found that AtbZIP16 is a negative regulator in light-mediated inhibition of cell elongation but is a positive regulator in light-regulated seed germination. AtbZIP16 negatively regulates and AtbZIP68 and AtGBF1 positively regulate the expression of light-harvesting chlorophyll a/b-binding protein (LHCB2.4) (Shen et al. 2008; Shaikhali et al. 2012). In addition, GBF1-interacting protein 1 (GIP1) functions to promote hypocotyl elongation during the early stages of A. thaliana seedling development by regulating the repression effect by bZIP16 and the activation effect by bZIP68 and GBF1 on LHCB2.4 expression (Shaikhali et al. 2015). The environmentally induced changes in the redox state regulate the activity of members of the G-group of bZIP TFs (Shaikhali et al. 2012). Moreover, GBF1 interacts with the bHLH master regulator MYC2 and the CAM7-TF in Arabidopsis. GBF1 physically interacts with MYC2 and inhibits each other through non-DNA binding heterodimers (Maurya et al. 2015), and important functions of GBF1 have been established in promoting lateral root development and natural senescence (Dröge-Laser et al. 2018). GBF from Catharanthus roseus, CrGBF1 and CrGBF2 (66.67% identity with BcbZIP134) play negative regulatory roles in the biosynthesis of terpenoid indole alkaloids (TIAs). Both CrGBF1 and CrGBF2 regulate the expression of strictosidine synthase gene (Str) via direct interaction with the G-box in the promoter of Str (Sibéril et al. 2001). These studies demonstrate the complexity of the regulatory network in functioning bZIP TF G-group members. A recent study showed that repressors CrGBFs dimerize with activator CrMYC2, and CrGBF1 binds to the same cis-elements (T/G-box) as CrMYC2 in the target gene promoters, suggesting that CrGBFs antagonize CrMYC2 transactivation possibly by competitive binding to the T/G-box in the target promoters and/or protein–protein interaction that forms a non-DNA binding complex that prevents CrMYC2 from binding to its target promoters, thus fine-tune the accumulation of TIAs (Sui et al. 2018). Earlier studies have shown that the DNA binding activity of parsley CPRF4a, with which BcbZIP134 shares the highest sequence identity (80.48%), is modulated in a phosphorylation-dependent manner and that cytosolic components are involved in the regulation of this process (Wellmer et al. 2001). Therefore, much effort is still needed to explore how BcbZIP134 performs its regulatory functions in B. chinense and whether it functions as repressor of SS biosynthesis through competing the binding site in promoters of enzymatic genes with some activators. In the model reported here, we have determined that BcbZIP134 overexpression led to the negative regulation of SS biosynthesis.
Conclusions
BcbZIP134, isolated from transcriptome of B. chinense, encodes a TF transcription factor belonging to bZIP G subfamily. MeJA can induce the high expression of BcbZIP134 in adventitious roots of B. chinense. Hairy roots of B. chinense overexpressing BcbZIP134 biosynthesize significantly less content of the main SS monomers a, c and d when compared to control hairy roots. The contents of a few other minor SS monomers such as acetyl-SS a, acetyl-SS d and unknown compounds were also decreased on different extent. It is revealed that BcbZIP134 negatively regulates the biosynthesis of saikosaponin in B. chinense.
References
Aoyagi H, Kobayashi Y, Yamada K, Yokoyama M, Kusakari K, Tanaka H (2001) Efficient production of saikosaponins in Bupleurum falcatum root fragments combined with signal transducers. Appl Microbiol Biotechnol 57:482–488. https://doi.org/10.1007/s002530100819
Ashour ML, Wink M (2011) Genus Bupleurum: a review of its phytochemistry, pharmacology and modes of action. J Pharm Pharmacol 63:305–321. https://doi.org/10.1111/j.2042-7158.2010.01170.x
Bathoju G, Rao K, Giri A (2017) Production of sapogenins (stigmasterol and hecogenin) from genetically transformed hairy root cultures of Chlorophytum borivilianum (Safed musli). Plant Cell Tissue Organ Cult 131:369–376. https://doi.org/10.1007/s11240-017-1290-8
Chen LR, Chen YJ, Lee CY, Lin TY (2007) MeJA-induced transcriptional changes in adventitious roots of Bupleurum kaoi. Plant Sci 173:12–24. https://doi.org/10.1016/j.plantsci.2007.03.013
Dong LM, Sui C, Liu YJ, Yang Y, Wei JH, Yang YF (2011) Validation and application of reference genes for quantitative gene expression analyses in various tissues of Bupleurum chinense. Mol Biol Rep 38:5017–5023. https://doi.org/10.1007/s11033-010-0648-3
Dröge-Laser W, Snoek BL, Snel B, Weiste C (2018) The arabidopsis, bZIP transcription factor family—an update. Curr Opin Plant Biol 45:36–49. https://doi.org/10.1016/j.pbi.2018.05.001
Fang W, Yang YJ, Guo BL, Cen S (2017) Anti-influenza triterpenoid saponins (saikosaponins) from the roots of Bupleurum marginatum var. stenophyllum. Bioorg Med Chem Lett 27:1654–1659. https://doi.org/10.1016/j.bmcl.2017.03.015
Gao K, Wu SR, Wang L, Xu YH, Wei JH, Sui C (2015) Cloning and analysis of β-amyrin synthase gene in Bupleurum chinense. Genes Genom 37:767–774. https://doi.org/10.1007/s13258-015-0307-0
Hurst HC (1995) Transcription factors 1: bZIP proteins. Protein Profile 2:101–168
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. https://doi.org/10.1016/S1360-1385(01)02223-3
Kim YS, Cho JH, Ahn J, Hwang B (2006) Upregulation of isoprenoid pathway genes during enhanced saikosaponin biosynthesis in the hairy roots of Bupleurum falcatum. Mol Cells 22:269–274. https://doi.org/10.1016/j.tcm.2006.07.002
Kim YS, Cho JH, Park S, Han JY, Back K, Choi YE (2011) Gene regulation patterns in triterpene biosynthetic pathway driven by overexpression of squalene synthase and methyl jasmonate elicitation in Bupleurum falcatum. Planta 233:343–355. https://doi.org/10.1007/s00425-010-1292-9
Kusakari K, Yokoyama M, Inomata S (2000) Enhanced production of saikosaponins by root culture of Bupleurum falcatum L. using two-step control of sugar concentration. Plant Cell Rep 19:1115–1120. https://doi.org/10.1007/s002990000240
Lee H, Kim KU, Son TK, Lee SC (2002) Effects of planting date and density on growth characteristics and saikosaponins content in bupleurum falcatum. Korean J Med Crop Sci 10:317–326
Li DQ, Wu J, Liu LY, Wu YY, Li LZ, Huang XX, Liu QB, Yang JY, Song SJ, Wu CF (2015) Cytotoxic triterpenoid glycosides (saikosaponins) from the roots of Bupleurum chinense. Bioorg. Med Chem Lett 25:3887–3892. https://doi.org/10.1016/j.bmcl.2015.07.053
Lin TY, Chiou CY, Chiou SJ (2013) Putative genes involved in saikosaponin biosynthesis in Bupleurum species. Int J Mol Sci 14:12806–12826. https://doi.org/10.3390/ijms140612806
Llorca CM, Potschin M, Zentgraf U (2014) bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci 5:169–183. https://doi.org/10.3389/fpls.2014.00169
Ma DM, Pu GB, Lei CY, Ma LQ, Wang HH, Guo YW, Chen JL, Du ZG, Wang H, Li GF, Ye HC, Liu BY (2009) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol 50:2146–2161. https://doi.org/10.1093/pcp/pcp149
Maurya JP, Sethi V, Gangappa SN, Gupta N, Chattopadhyay S (2015) Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J 83:439–450. https://doi.org/10.1111/tpj.12899
Memelink J, Gantet P (2007) Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem Rev 6:353–362. https://doi.org/10.1007/s11101-006-9051-z
Mertens J, Pollier J, Vanden Bossche R, Lopez-Vidriero I, Franco-Zorrilla JM, Goossens A (2016) The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol 170:194–210. https://doi.org/10.1104/pp.15.01645
Miyamoto K, Nishizawa Y, Minami E, Nojiri H, Yamane H, Okada K (2014) Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J Plant Physiol 173:19–27. https://doi.org/10.1016/j.jplph.2014.09.001
Moses T, Pollier J, Almagro L, Buyst D, Van Montagu M, Pedreño MA, Martins JC, Thevelein JM, Goossens A (2014) Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc Natl Acad Sci USA 111:1634–1639. https://doi.org/10.1073/pnas.1323369111
Nasrollahi V, Mirzaie-Asl A, Piri K, Nazeri S, Mehrabi R (2014) The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 103:32–37. https://doi.org/10.1016/j.phytochem.2014.03.004
Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H (2009) OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J Biol Chem 284:26510–26518. https://doi.org/10.1016/j.phytochem.2014.03.004
Selmar D (2008) Potential of salt and drought stress to increase pharmaceutical significant secondary compounds in plants. Landbauforsch Volk 58:139–144.https://doi.org/10.1074/jbc.M109.036871
Shaikhali J (2015) GIP1 protein is a novel cofactor that regulates DNA-binding affinity of redox-regulated members of bZIP transcription factors involved in the early stages of Arabidopsis development. Protoplasma 252:867–883. https://doi.org/10.1007/s00709-014-0726-9
Shaikhali J, Norén L, de Dios Barajas-López J, Srivastava V, König J, Sauer UH, Wingsle G, Dietz KJ, Strand A (2012) Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in arabidopsis. J Biol Chem 287:27510–27525. https://doi.org/10.1074/jbc.M112.361394
Shen H, Cao K, Wang X (2008) AtbZIP16 and AtbZIP68, two new members of GBFs, can interact with other G group bZIPs in Arabidopsis thaliana. BMB Rep 41:132–138. https://doi.org/10.5483/BMBRep.2008.41.2.132
Sibéril Y, Benhamron S, Memelink J, Giglioli-Guivarc’h N, Thiersault M, Boisson B, Doireau P, Gantet P (2001) Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol Biol 45:477–488. https://doi.org/10.1023/A:1010650906695
Sui C, Zhang J, Wei JH, Chen SL, Li Y, Xu JS, Jin Y, Xie CX, Gao ZH, Chen HQ, Yang CM, Zhang Z, Xu YH (2011) Transcriptome analysis of Bupleurum chinense focusing on genes involved in the biosynthesis of saikosaponins. BMC Genom 12:539. https://doi.org/10.1186/1471-2164-12-539
Sui C, Chen M, Xu JS, Wei JH, Jin Y, Xu YH, Sun J, Gao K, Yang CM, Zhang Z, Chen SL, Luo HM (2014) Comparison of root transcriptomes and expressions of genes involved in main medicinal secondary metabolites from Bupleurum chinense and Bupleurum scorzonerifolium, the two Chinese official Radix bupleuri source species. Physiol Plant 153:230–242. https://doi.org/10.1111/ppl.12254
Sui X, Singh SK, Patra B, Schluttenhofer C, Guo W, Pattanaik S, Yuan L (2018) Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J Exp Bot 69:4267–4281. https://doi.org/10.1093/jxb/ery229
Tamura K, Yoshida K, Hiraoka Y, Sakaguchi D, Chikugo A, Mochida K, Kojoma M, Mitsuda N, Saito K, Muranaka T, Seki H (2018) The basic helix-loop-helix transcription factor GubHLH3 positively regulates soyasaponin biosynthetic genes in glycyrrhiza uralensis. Plant Cell Physiol 59:778–791. https://doi.org/10.1093/pcp/pcy046
Wellmer F, Schäfer E, Harter K (2001) The DNA binding properties of the parsley bZIP transcription factor CPRF4a are regulated by light. J Biol Chem 276:6274–6279. https://doi.org/10.1074/jbc.M007971200
Xu Z, Song J (2017) The 2-oxoglutarate-dependent dioxygenase superfamily participates in tanshinone production in Salvia miltiorrhiza. J Exp Bot 68:2299–2308. https://doi.org/10.1093/jxb/erx113
Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5:353–365. https://doi.org/10.1093/mp/ssr087
Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, Gantet P, Memelink J (2011a) The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate responsive expression of the ORCA genes regulating alkaloid biosynthesis in Catharanthus roseus. Plant J 67:61–71. https://doi.org/10.1111/j.1365-313X.2011.04575.x
Zhang Y, Zheng S, Liu Z, Wang L, Bi Y (2011b) Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol 168:367–374. https://doi.org/10.1016/j.jplph.2010.07.025
Zhang FY, Fu XQ, Lv ZY, Lu XL, Shen Q, Zhang L, Zhu MM, Wang GF, Sun XF, Liao ZH, Tang KX (2015a) A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol Plant 8:163–175. https://doi.org/10.1016/j.molp.2014.12.004
Zhang R, Zhang BL, Li GC, Xie T, Hu T, Luo ZY (2015b) Enhancement of ginsenoside Rg1 in Panax ginseng hairy root by overexpressing the ɑ-L-rhamnosidase gene from Bifidobacterium breve. Biotechnol Lett 37:2091–2096. https://doi.org/10.1007/s10529-015-1889-y
Zhang Y, Zhou ZY, Xia PG, Liang ZS, Liu SL, Liu ZL (2016) Expression of key enzyme genes and content of saikosaponin in saikosaponin biosynthesis under drought stress in bupleurum chinense. China J Chin Mater Med 41:643–647. https://doi.org/10.4268/cjcmm20160416
Zhao Q, Cui MY, Levsh O, Yang DF, Liu J, Li J, Hill L, Yang L, Hu YH, Weng JK, Chen XY, Martin C (2018) Two CYP82D enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4′-deoxyflavones in Scutellaria baicalensis. Mol Plant 11:135–148. https://doi.org/10.1016/j.molp.2017.08.009
Zhu Z, Liang Z, Han R, Xin W (2009a) Impact of fertilization on drought response in the medicinal herb bupleurum chinense DC.: growth and saikosaponin production. Ind Crop Prod 29:629–633. https://doi.org/10.1016/j.indcrop.2008.08.002
Zhu ZB, Liang ZS, Han RL (2009b) Saikosaponin accumulation and antioxidative protection in drought-stressed Bupleurum chinense DC. plants. Environ Exp Bot 66:326–333. https://doi.org/10.1016/j.envexpbot.2009.03.017
Zhu L, Liang ZT, Yi T, Ma Y, Zhao ZZ, Guo BL, Zhang JY, Chen HB (2017) Comparison of chemical profiles between the root and aerial parts from three Bupleurum species based on a UHPLC-QTOF-MS metabolomics approach. BMC Complem Altern Med 17:305. https://doi.org/10.1186/s12906-017-1816-y
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 81773847), the CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant No. 2016-I2M-2-003), the National Standardization Project of Traditional Chinese Medicine (Grant No. ZYBZH-Y-JIN-34) and the Sichuan Province “13th Five-Year” Crop and Livestock Breeding Key Varieties Breeding Projects (Grant No. 2016NYZ0020). The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Author information
Authors and Affiliations
Contributions
CS and JW: Conceptualization; JX, SW, YX and ZG: Data curation; JX and SW: Investigation; JX and SW: Methodology; CS and JW: Project administration; SW: Writing—original draft; CS: Writing—review & editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Ali R. Alan.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Primers used in the present study
Appendix: Primers used in the present study
Purposes | Names | Sequences (5′–3′) |
|---|---|---|
qRT-PCR for β-Tubulin | Tu-F | ATGTTCAGGCGCAAGGCTT |
Tu-R | TCTGCAACCGGGTCATTCAT | |
qRT-PCR for Actin | At-F | TCACCATTGGAGCTGAGAGATTC |
At-R | CTGCAGCTTCCATTCCAATCA | |
qRT-PCR for BcbZIP134 | bZIP134-F | CCTCCATTAGTGGCTGTTCC |
bZIP134-R | CTTATCCATTCAGTCCATTTGC | |
3′-RACE for BcbZIP134 | GSP3 | GCGGTGAAGTGGATGGTAAGGGGTCTGA |
NGSP3 | TGGATGGTAAGGGGTCTGAGGGGAAGGA | |
5′-RACE for BcbZIP134 | GSP5 | TCCTTCCCCTCAGACCCCTTACCATCCA |
NGSP5 | CCCCTTACCATCCACTTCACCGCTTCC | |
Full-length cloning PCR for BcbZIP134 | FL134-F | TGTTCGAGTTGAGTTAGAAGCTACAAT |
FL134-R | GAACACCCATCTAAATGCATACAAG | |
Construction of overexpression vectors for BcbZIP134 | OE134-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTGATGGGTAGTA GTGATATGGA AAAATC |
OE134-R | GGGGACCACTTTGTACAAGAAAGCTGGGTGTCAGAGACCAGTTTGTGCGGGCTCTC | |
Verification for BcbZIP134 insertion | P35S | GACGCACAATCCCACTATCC |
T35S | GCTCAACACATGAGCGAAAC | |
qRT-PCR for HMGR | HMGR-F | TATAACTGGCGATGTGGTGAAG |
HMGR-R | GTGGCTATGAAGATTGCAGAAAC | |
qRT-PCR for FPS | FPS-F | CTCACACACGCAGAGGTCAA |
FPS-R | TGGGGATATGGTTGCGAAG | |
qRT-PCR for IPPI | IPPI-F | AGGTGACATTCCCTTTGGTG |
IPPI-R | AGAAGCTTCCTCTGTGCAGC | |
qRT-PCR for SQS | SQS-F | AGGTTACCAGGAAGCAATAGAA |
SQS-R | CGACAACCCTAATCCAACAAG | |
qRT-PCR for SQE | SQE-F | GGTGGAGGAATGACGGTTGC |
SQE-R | TACGGGCTTGCGAAGGGTGT | |
qRT-PCR for β-AS | AS-F | ACATGGCTTTCGATACTCGG |
AS-R | ATTTTCGCTGGATGCATAGG |
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Xu, J., Wu, SR., Xu, YH. et al. Overexpression of BcbZIP134 negatively regulates the biosynthesis of saikosaponins. Plant Cell Tiss Organ Cult 137, 297–308 (2019). https://doi.org/10.1007/s11240-019-01571-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01571-0