Abstract
Plants produce alkaloids, among others, to protect themselves against microbial infection, herbivore attack or ultraviolet irradiation. For man, alkaloid metabolism is the source of many natural products with useful applications, including pharmaceuticals. A major mechanism regulating alkaloid production in plant cells is the control of the transcription of the biosynthetic genes. Several transcription factors involved in the regulation of alkaloid biosynthesis genes have been isolated and studied. There are indications that the abundance and activities of transcription factors themselves are regulated by external signals. The aim of this review is to give an update on the transcriptional regulation of terpenoid indole metabolism in Catharanthus roseus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most classes of secondary metabolites are accumulated by plants in response to certain developmental or environmental signals in specific organs or cell types. The anthocyanin pigments for example, accumulate specifically in certain cell types of the flower petal in many plant species. However, other parts of the plant are able to produce anthocyanins in response to various stress conditions, such as excessive UV light. During the past two decades, it has emerged that these specific patterns of accumulation are largely, although not exclusively, due to specific transcriptional regulation of the genes encoding biosynthesis enzymes and metabolite transporters. It turns out that these genes are regulated in a coordinate manner in response to developmental and environmental signals by specific transcription factors.
This contribution about the transcriptional regulation of the terpenoid indole alkaloid (TIA) pathway in Catharanthus roseus is an update of previous reviews (Meijer et al. 1993a; Memelink et al. 2001a, b; Hilliou et al. 2001; Gantet and Memelink 2002; Vom Endt et al. 2002; Pauw and Memelink 2005).
Regulation of the terpenoid indole alkaloid biosynthetic pathway
Research on the terpenoid indole alkaloids (TIAs) is mainly primed by the pharmaceutical applications of several of the compounds. The monomeric alkaloids serpentine and ajmalicine are used as tranquilizer and to reduce hypertension, respectively. The dimeric alkaloids vincristine and vinblastine are potent antitumor drugs. In plants, TIAs are thought to be involved in defence responses. Several reports show that physiological concentrations of TIAs can have antifeeding activity or can delay growth of insect larvae, fungi and microbes (Aerts et al. 1991, 1992; Luijendijk et al. 1996). Terpenoid indole alkaloids are found in a limited number of plant species belonging to the plant families Apocynaceae, Loganiaceae, Rubiaceae and Nyssaceae. The best progress on molecular characterization of the pathway has been made with Catharanthus roseus (L.) G. Don (Madagascar periwinkle), a member of the Apocynaceae family. C. roseus cells have the genetic potential to synthesize over a hundred terpenoid indole alkaloids.
The initial step in TIA biosynthesis is the condensation of tryptamine with the iridoid glucoside secologanin (Fig. 1). This condensation is performed by the enzyme strictosidine synthase (STR) and results in the synthesis of 3α(S)-strictosidine. Strictosidine is deglucosylated by strictosidine β-d-glucosidase (SGD). Further enzymatic and spontaneous conversions result in the biosynthesis of numerous TIAs. Tryptamine, providing the indole moiety of TIAs, is formed by decarboxylation of tryptophan by the enzyme tryptophan decarboxylase (TDC). Secologanin, providing the terpenoid part of the TIAs, is synthesized via multiple enzymatic conversions from geraniol. The terpenoid precursors that constitute the backbone of geraniol are produced via the MEP (2-C-methyl-d-erythritol 4-phosphate) pathway (Contin et al. 1998). Most steps involved in the conversion of geraniol to secologanin are unknown. Geraniol 10-hydroxylase (G10H) catalyses the first committed step in the formation of secologanin by 10-hydroxylation of geraniol, whereas secologanin synthase (SLS) catalyses the last conversion of loganin into secologanin. G10H and SLS are both cytochrome P450 monooxygenases, which require the co-enzyme NADPH:cytochrome P450 reductase (CPR). Dimeric alkaloids are formed by peroxidase-catalyzed condensation of vindoline and catharanthine (Sottomayor et al. 2004). Many monomeric TIAs are found in all plant organs, but vindoline and vindoline-derived dimeric alkaloids are only found in chloroplast-containing plant tissues. Vindoline is derived via a number of steps from tabersonine. The first step is catalyzed by the P450 enzyme tabersonine 16-hydroxylase (T16H), which also requires CPR as a co-enzyme. The two final steps are catalyzed by acetyl CoA:deacetylvindoline 4-O-acetyltransferase (DAT) and the 2-oxoglutarate-dependent dioxygenase desacetoxyvindoline-4-hydroxylase (D4H).
Biosynthetic pathway for terpenoid indole alkaloids in Catharanthus roseus. Solid arrows indicate single enzymatic conversions, whereas dashed arrows indicate multiple enzymatic conversions. Indicated are enzymes, for which the corresponding genes were cloned. Enzymes encoded by genes regulated by ORCA3 are shown against a black background. AS: anthranilate synthase, CPR: cytochrome P450 reductase, D4H: desacetoxyvindoline 4-hydroxylase, DAT: acetyl-CoA:4-O-deacetylvindoline 4-O-acetyltransferase, DXS:d-1-deoxyxylulose 5-phosphate synthase, G10H: geraniol 10-hydroxylase, PRX1: peroxidase 1, SGD: strictosidine β-d-glucosidase, SLS: secologanin synthase, STR: strictosidine synthase TDC: tryptophan decarboxylase, T16H: tabersonine 16-hydroxylase
The biosynthesis of TIAs is highly regulated, and depends on tissue- and cell-specific factors as well as environmental signals. Developmental control of TIA biosynthesis and gene expression has previously been reviewed (De Luca and St-Pierre 2000). Here, we will restrict ourselves to stress signals that regulate TIA biosynthesis and gene expression. Inducing effects on alkaloid metabolism were observed for fungal elicitors (Moreno et al. 1995), jasmonates (Aerts et al. 1994; Vazquez-Flota and De Luca 1998; Gantet et al. 1998; Rijhwani and Shanks 1998), auxin starvation (Gantet et al. 1998), UV-B light (Ouwerkerk et al. 1999a) and wounding (Vazquez-Flota et al. 2004).
cDNA clones encoding STR (McKnight et al. 1990; Pasquali et al. 1992) and genomic sequences (Pasquali et al. 1999) have been isolated. A TDC cDNA (De Luca et al. 1989) and corresponding genomic clone (Goddijn et al. 1994) have also been reported. STR and TDC mRNA accumulate in suspension-cultured cells after auxin starvation (Pasquali et al. 1992; Goddijn et al. 1992), and exposure to fungal elicitors (Pasquali et al. 1992; Roewer et al. 1992) or (methyl) jasmonate (Menke et al. 1999a). Elicitor-responsive expression of TDC and STR depends on jasmonates as a secondary signal (Menke et al. 1999a). In leaves TDC and STR are induced by a UV-B light pulse (Ouwerkerk et al. 1999a). Since TDC and STR are encoded by single-copy genes in C. roseus (Pasquali et al. 1992; Goddijn et al. 1994), all these signals act on the same promoter regions. In addition, cDNA clones for G10H (Collu et al. 2001), SLS (Irmler et al. 2000), CPR (Meijer et al. 1993b), which is essential for the G10H- and SLS-catalysed reactions, and SGD (Geerlings et al. 2000) have been isolated. CPR mRNA accumulation is rapidly induced by fungal elicitor (Meijer et al. 1993b) and the CPR promoter is elicitor-responsive in transgenic tobacco (Lopes Cardoso et al. 1997). All three genes are induced by MeJA in C. roseus cell cultures (Geerlings et al. 2000; van der Fits and Memelink 2000; Collu et al. 2001).
From the vindoline pathway, cDNA clones for T16H (Schröder et al. 1999), DAT (St-Pierre et al. 1998), and D4H (Vazquez-Flota et al. 1997) have been isolated. The expression of the corresponding genes is light-regulated in seedlings. In addition, D4H and DAT were shown to be induced by methyl-jasmonic acid (MeJA) in cell cultures (van der Fits and Memelink 2000).
Finally, for a vacuolar peroxidase, which can dimerize vindoline and catharanthine to form α-3′,4′-anhydrovinblastine (Sottomayor and Ros Barcelo 2003), the direct precursor of vinblastine, a cDNA clone was isolated (Sottomayor et al. 2004).
The observations that TDC and STR mRNAs coordinately accumulate in response to fungal elicitors, jasmonates, UV light, and auxin starvation, clearly indicate that the TDC and STR genes are controlled by common regulators. Since all TIA biosynthetic genes tested were induced by MeJA (van der Fits and Memelink 2000; Collu et al. 2001), this suggests that a common JA-responsive regulator controls many and possibly all TIA structural genes.
Promoter studies have identified a regulator of alkaloid biosynthesis
Due to lack of genetic tools for C. roseus and easily visible phenotypes, research aiming at isolating regulators of TIA biosynthesis genes focused on the corresponding promoter sequences. Promoter analysis of the STR and TDC genes revealed that both contained sequences involved in the regulation by stress signals such as UV-irradiation and fungal elicitors (Ouwerkerk et al. 1999a, b; Ouwerkerk and Memelink 1999b; Pasquali et al. 1999). The plant stress hormone jasmonic acid is an essential second messenger for elicitor-induced STR and TDC gene expression (Menke et al. 1999a).
A short STR promoter region called RV, which contains a JA- and elicitor-responsive element (JERE) is responsible for JA- and elicitor-responsive gene expression (Menke et al. 1999b) (Fig. 2). Using the RV region as bait in yeast one-hybrid screening, a cDNA encoding ORCA2 (Octadecanoid-Responsive Catharanthus AP2-domain protein 2) was isolated. ORCA2 is a transcription factor of the AP2 (APETALA2)-domain family, which is characterized by the DNA-binding AP2-domain. ORCA2 mRNA accumulation is rapidly induced by MeJA. In transient assays ORCA2 activates STR gene expression by interacting with the JERE (Menke et al. 1999b). These data indicate that ORCA2 controls the JA-responsive expression of the STR gene and possibly other TIA biosynthesis genes.
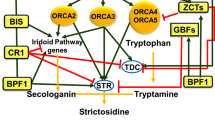
Overview of transcription factors that can interact with the STR and TDC promoters. Perception of an elicitor from yeast extract (YE) leads to an increase in JA levels, which is necessary for the activation of the ORCA transcription factors. Although the cellular location of the YE receptor is unknown, it is tentatively placed in the plasma membrane. The ORCA transcription factors can activate gene expression via interaction with the TDC promoter and the RV fragment of the STR promoter. Although the ORCA binding site in the TDC promoter has not been precisely mapped, it is tentatively indicated downstream of the DB fragment. In addition, YE rapidly induces accumulation of mRNAs encoding ZCT proteins, which can repress gene expression via binding to the DB fragment of the TDC promoter and the BA and, to a lesser extent, the RV fragments of the STR promoter. Also, YE induces accumulation of mRNA encoding CrBPF1, which is putatively involved in regulation of STR via interaction with the BA region. CrGBF transcription factors can repress STR promoter activity via binding to a G-box in the NR region. YE, yeast elicitor; JA, jasmonic acid; BPF-1, Catharanthus roseus box P-binding factor 1 homologue; GBF, G-box-binding factor; STR, strictosidine synthase; ZCT, zinc-finger Catharanthus transcription factor; ORCA, octadecanoid-responsive Catharanthus AP2-domain protein; TDC, tryptophan decarboxylase
ORCA3, a central regulator of alkaloid biosynthesis
A closely related TIA regulatory gene, called ORCA3, was isolated via a T-DNA activation tagging approach applied to a C. roseus cell culture (van der Fits and Memelink 2000; van der Fits et al. 2001). ORCA3 also binds to the JERE element and activates STR gene expression in transient assays (van der Fits and Memelink 2001).
ORCA3 gene expression is also induced by MeJA, indicating that ORCA3 may have overlapping functions with ORCA2 in regulating JA-responsive expression of alkaloid biosynthesis genes. Overexpression of ORCA3 in C. roseus cultured cells increased the expression of the TIA biosynthesis genes TDC, STR, CPR, D4H (van der Fits and Memelink 2000) and SLS (unpublished results; Fig. 1). Interestingly, ORCA3 also regulated two genes encoding enzymes of primary metabolism leading to TIA precursor formation: ASα (α subunit of anthranilate synthase) and DXS (D-1-deoxyxylulose 5-phosphate synthase). This suggested that ORCA3 is a central regulator of TIA biosynthesis, acting on several steps of the TIA pathway and also regulating the biosynthesis of TIA precursors. However, ORCA3 was not found to regulate G10H and DAT. Transgenic cells overexpressing ORCA3 accumulated significantly more tryptophan and tryptamine, but no TIAs were detected, suggesting that the terpenoid branch of the pathway remained limiting for TIA production. This was confirmed by the fact that when the cells were fed with the terpenoid precursor loganin, ORCA3 overexpression caused an increase in TIA production (van der Fits and Memelink 2000).
How do the ORCA proteins mediate JA-responsive expression of TIA biosynthesis genes?
ORCA mRNA accumulation is rapidly and transiently induced by MeJA and the peaks of induced ORCA expression preceded the maximal induction of the target genes TDC and STR (Menke et al. 1999b; van der Fits and Memelink 2001). This suggests that JA induces an increase in the amount of ORCA proteins, which may be sufficient to activate the expression of the target genes. However, expression studies with the protein synthesis inhibitor cycloheximide (CHX) showed that MeJA induces the expression of the STR and TDC genes in a CHX-insensitive manner (van der Fits and Memelink 2001), demonstrating that de novo synthesis of ORCA transcription factors is not necessary. Therefore, MeJA does not induce TIA gene expression simply by increasing ORCA protein abundance, but instead appears to activate pre-existing ORCA protein. This may occur via post-translational modifications and/or protein–protein interactions (Vom Endt et al. 2002). One of the possible post-translational modifications is phosphorylation. Reversible phosphorylation is a prevalent mechanism by which the activity of eukaryotic transcription factors is regulated in response to changes in the cellular environment (Holmberg et al. 2002). The induction of STR expression by JA is sensitive to protein kinase inhibitors (Menke et al. 1999a), which is compatible with the possibility that the ORCA proteins are phosphorylated, although any protein in the signal transduction pathway leading to ORCA activation could be a phosphoprotein. In addition to activating ORCA proteins, JA also activates an upstream transcription factor distinct from the ORCAs, which then induces ORCA3 gene expression via binding to a JA-responsive element in its promoter (Fig. 3; Vom Endt 2004). This JA-responsive element in the ORCA3 promoter is different from the JERE in the STR promoter. This indicates that JA switches on a small transcriptional cascade consisting of this unknown upstream transcription factor and ORCA3. However, this transcriptional cascade is not necessary for JA-responsive expression of TIA biosynthesis genes (Fig. 3).
Model for JA-induced STR gene expression mediated by the ORCA3 transcription factor in Catharanthus roseus. JA directly activates the ORCA3 protein post-translationally, indicated by an asterisk. Active ORCA3 stimulates STR gene expression by binding to the JA- and elicitor-responsive element (JERE). JA also post-translationally activates an unknown upstream transcription factor, which then induces ORCA3 gene expression via binding to a JA-responsive element (JRE). ORCA3, Octadecanoid-Responsive Catharanthus AP2-domain protein 3; TAF, transcription activating factor; STR, strictosidine synthase
Additional transcription factors may control alkaloid biosynthesis
Although ORCA3 plays an important role in regulating TIA biosynthesis, it is not sufficient by itself to regulate the complete pathway. This suggests that one or more other transcription factors have complementary gene targets. For example it would be interesting to know the target genes for ORCA2.
The use of another JA- and elicitor-responsive region (BA) of the STR promoter, which is distinct from the RV region (Fig. 2), as bait in a yeast one-hybrid screen resulted in the isolation of a periwinkle homologue of the MYB-like transcription factor BPF1 from parsley (CrBPF1) (van der Fits et al. 2000). CrBPF1 mRNA accumulation is induced by elicitor but not by MeJA, suggesting that elicitor induces STR gene expression in periwinkle cells via JA-dependent and independent pathways.
In addition, the STR promoter contains a conserved plant promoter element called G-box (5′-CACGTG-3′) located in the NR region immediately upstream of the JERE element in the RV region (Fig. 2). This G-box was shown to be an active cis-regulatory element in planta (Ouwerkerk and Memelink 1999a). A yeast one-hybrid screen using the G-box as bait allowed isolation of G-box binding factors (CrGBF) of the basic leucine zipper (bZIP) class and MYC-type basic helix-loop-helix (bHLH) (CrMYC) transcription factors (Pré et al. 2000; Chatel et al. 2003). In transient assays CrGBF1 and CrGBF2 were shown to be repressors of STR gene expression (Sibéril et al. 2001). The CrGBF transcription factors were also able to bind in vitro to a G-box like element in the TDC promoter (Sibéril et al. 2001) suggesting that CrGBFs could coordinately regulate several TIA biosynthesis genes. MeJA induces the accumulation of an mRNA encoding the (bHLH) transcription factor CrMYC1, which binds specifically to the G-box element in yeast (Chatel et al. 2003). Whether CrMYC1 has an effect on STR gene expression has not been tested. Interestingly, G-box and G-box-like elements were found to be essential for the JA-responsiveness of several promoters (Pauw and Memelink 2005). In Arabidopsis, the bHLH transcription factor AtMYC2 (also called JIN1 or RAP-1), which binds to the G-box (de Pater et al. 1997), regulates the expression of certain JA-responsive genes (Lorenzo et al. 2004; Boter et al. 2004). Two homologous bHLH transcription factors from tomato, JAMYC2 and JAMYC10, bind to a G-box like sequence in the JA-responsive promoter of the leucine aminopeptidase gene (Boter et al. 2004). The expression of the AtMYC2 gene and the JAMYC genes is induced by JA. Several of the AtMYC2 target genes were shown to be secondary JA-responsive genes, since CHX abolished their JA-responsive expression (Rojo et al. 1998; Jensen et al. 2002). In this respect, the regulation of JA-responsive gene expression by AtMYC2 is clearly different from ORCA-mediated expression of TIA biosynthesis genes in response to JA. Apparently, the bHLH factors are synthesized de novo in response to JA, and in this manner confer the JA-responsive expression of these secondary response genes via binding to G-box-like sequences in their promoters.
Yeast one-hybrid screening with the elicitor-responsive DB element of the TDC promoter led to the isolation of three members of the Cys2/His2-type zinc finger gene family in C. roseus, encoding ZCT1, ZCT2, and ZCT3 (Pauw et al. 2004). The ZCT proteins contain two zinc fingers and belong to the EPF subfamily of TFIIIA-type zinc finger proteins in plants. Members of this subfamily are characterized by the highly conserved sequence QALGGH in their zinc finger motifs, which is essential for DNA binding. Recombinant ZCT proteins bind in a zinc-dependent manner to the TDC promoter as well as the STR promoter. In transient trans-activation assays, the ZCT proteins can repress the activity of these promoters. All three ZCT proteins contain the LxLxL motif, which has been demonstrated in other zinc finger transcription factors, including proteins that are highly similar in amino acid sequence to the ZCTs, to be involved in active repression (Ohta et al. 2001; Hiratsu et al. 2003). It seems therefore likely that this LxLxL motif is responsible for the repressor activity of the ZCT proteins. Within the natural STR promoter context, the ZCT proteins can repress the activating activity of the ORCAs without competing for the same binding sites. ZCT mRNA levels were increased by yeast elicitor and MeJA. Yeast elicitor induced ZCT gene expression after 30 min (Pauw et al. 2004) and JA biosynthesis after 2 h (Menke et al. 1999a). Therefore, the induction of ZCT gene expression by yeast elicitor seems to be upstream or independent of the induction of JA biosynthesis. The fact that the ZCT repressors can bind to elicitor- and JA-responsive regions of the STR and TDC promoters, and the fact that ZCT expression levels were induced by elicitor and MeJA treatment, indicates that these proteins are involved in regulation of TDC and STR expression by elicitor and JA. There are several mechanisms by which the ZCT proteins could actively repress transcription of the STR and TDC promoters. The ZCT proteins could prevent the association of a transcriptional activator with these promoters or could suppress the function of a DNA-bound transcriptional activator protein. Alternatively, ZCT proteins could have negative effects on the basal transcription machinery or could induce the formation of an inactive chromatin structure at the sites of the STR and TDC promoters.
Finally, it needs to be said that for all these transcription factors except the ORCA proteins, the involvement in regulating TIA biosynthesis genes has not been rigorously proven. For the ORCA transcription factors, it has been shown that STR promoter mutations that abolish in vitro binding, also abolish JA-responsive promoter activity in vivo (Menke et al. 1999b; van der Fits and Memelink 2001). In addition, it has been shown that overexpression of ORCA3 (van der Fits and Memelink 2000) or ORCA2 (unpublished results) leads to elevated expression levels of STR and other TIA biosynthesis genes. Such studies have not been performed for any of the other transcription factors. The only indication that they might be involved in regulating TIA biosynthesis genes is their binding to the STR and/or TDC promoters in vitro or in yeast, but whether they really do so in planta remains to be demonstrated.
Concluding remarks
Because of their pleiotropic action on a wide array of genes involved in metabolic differentiation of plant cells, central transcription factors enable the development of new strategies to engineer complex metabolic pathways and hold great promise for increasing the levels of pharmaceutically active molecules in plant cells for industrial production. This is of great interest if we consider that about 25% of all contemporary medicines and 50% of current anti-cancer drugs are derived from plants. In addition, plant secondary metabolites represent an enormous chemical diversity with largely unexplored pharmacological activities.
The challenge is therefore to find such central regulators of secondary metabolism. For terpenoid indole alkaloid metabolism in Catharanthus roseus, recent research has uncovered a number of candidate regulators. The studies have focused on transcription factors involved in elicitor- and JA-responsive gene expression, since these signaling compounds are known to have a stimulatory effect on alkaloid biosynthesis (Memelink et al. 2001a).
The following picture has emerged from transcription factor studies in C. roseus. Perception of an elicitor from yeast extract activates the octadecanoid pathway, which leads to an increase in JA levels. JA induces expression of the ORCA genes, and activates pre-existing ORCA proteins post-translationally (Fig. 3). The ORCA proteins can activate gene expression via interaction with the TDC promoter and the elicitor- and JA-responsive RV fragment of the STR promoter. In addition, elicitor rapidly induces the expression of the zinc finger proteins, which can repress gene expression via binding to the elicitor-responsive DB fragment of the TDC promoter and the elicitor- and JA-responsive BA and RV fragments of the STR promoter (Fig. 2). Also, elicitor induces accumulation of mRNA encoding CrBPF1, which is putatively involved in regulation of STR via interaction with the BA region. CrGBF transcription factors can repress STR promoter activity via binding to the G-box in the NR region (Fig. 2). This G-box can also interact with the bHLH transcription factor CrMYC1 (not shown in Fig. 2).
For all these transcription factors except the ORCA proteins, it needs to be demonstrated that they really regulate TIA biosynthesis genes in planta. If they do, it raises the question about the functional importance of the induction of both activators (ORCA) and repressors (CrGBF and ZCT) of STR and TDC gene expression by elicitor and JA. The simultaneous induction of repressors and activators may serve to fine-tune the amplitude and timing of gene expression. Such a fine-tuning may in part be achieved by the differential effect of elicitor and (Me)JA on the amplitude and kinetics of ORCA and ZCT mRNA accumulation (Pauw et al. 2004). Alternatively, in analogy to models used to explain switch-like transcriptional control by developmental signals (Barolo and Posakony 2002), induction of a combination of activators and repressors may be necessary to achieve a switch-like on/off state of gene expression in response to stress signals.
Many enzymatic steps in the TIA pathway are still unknown. The use of central transcriptional regulators, such as the ORCA proteins, for plant metabolic engineering might avoid the time-consuming step of acquiring knowledge about all enzymatic steps of this complex metabolic pathway (Gantet and Memelink 2002). On the other hand, it may require the equally time-consuming step of unraveling the equally complex regulatory network.
Abbreviations
- AP2:
-
APETALA2
- bHLH:
-
Basic helix-loop-helix
- bZIP:
-
Basic leucine zipper
- CrBPF-1:
-
C. roseus box P-binding factor 1 homologue
- CrGBF:
-
C. roseus G-box binding factor
- JA:
-
Jasmonic acid
- JERE:
-
Jasmonate- and elicitor-responsive element
- MeJA:
-
Methyl-jasmonate
- ORCA:
-
Octadecanoid-responsive Catharanthus AP2-domain
- STR:
-
Strictosidine synthase
- TDC:
-
Tryptophan decarboxylase
- TIA:
-
Terpenoid indole alkaloid
- ZCT:
-
Zinc-finger Catharanthus transcription factor
References
Aerts RJ, Snoeijer W, Aerts-Teerlink O, van der Meijden E, Verpoorte R (1991) Control and biological implications of alkaloid synthesis in Cinchona seedlings. Phytochemistry 30:3571–3577
Aerts RJ, Stoker A, Beishuizen M, van de Heuvel M, van der Meijden E, Verpoorte R (1992) Detrimental effects of Cinchona leaf alkaloids on larvae of the polyphagous insect Spodoptera exigua. J Chem Ecol 18:1955–1964
Aerts RJ, Gisi D, De Carolis E, De Luca V, Baumann TW (1994) Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. Plant J 5:635–643
Barolo S, Posakony JW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16:1167–1181
Boter M, Ruiz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18:1577–1591
Chatel G, Montiel G, Pré M, Memelink J, Thiersault M, Saint-Pierre B, Doireau P, Gantet P (2003) CrMYC1, a Catharanthus roseus elicitor-and jasmonate-responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J Exp Bot 54:2587–2588
Collu G, Unver N, Peltenburg-Looman AMG, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508:215–220
Contin A, van der Heijden R, Lefeber AWM, Verpoorte R (1998) The iridoid glucoside secologanin is derived from the novel triose phosphate/pyruvate pathway in Catharanthus roseus cell culture. FEBS Lett 434:413–416
De Luca V, Marineau C, Brisson N (1989) Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: comparison with animal dopa decarboxylases. Proc Natl Acad Sci USA 86:2582–2586
De Luca V, St Pierre B (2000) The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci 5:168–173
de Pater S, Pham K, Memelink J, Kijne J (1997) RAP-1 is an Arabidopsis MYC-like R protein homologue, that binds to G-box sequence motifs. Plant Mol Biol 34:169–174
Gantet P, Imbault N, Thiersault M, Doireau P (1998) Necessity of a functional octadecanoic pathway for indole alkaloid synthesis by Catharanthus roseus cell suspensions cultured in an auxin-starved medium. Plant Cell Physiol 39:220–225
Gantet P, Memelink J (2002) Transcription factors: tools to engineer the production of pharmacologically active plant metabolites. Trends Pharmacol Sci 23:563–569
Geerlings A, Martinez-Lozano Ibanez M, Memelink J, van der Heijden R, Verpoorte R (2000) Molecular cloning and analysis of strictosidine β-d-glucosidase, an enzyme in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J Biol Chem 275:3051–3056
Goddijn OJM, de Kam RJ, Zanetti A, Schilperoort RA, Hoge JHC (1992) Auxin rapidly down-regulates transcription of the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol 18:1113–1120
Goddijn OJM, Lohman FP, de Kam RJ, Schilperoort RA, Hoge JHC (1994) Nucleotide sequence of the tryptophan decarboxylase gene of Catharanthus roseus and expression of tdc-gusA gene fusions in Nicotiana tabacum. Mol Gen Genet 242:217–225
Hilliou F, van der Fits L, Memelink J (2001) Molecular regulation of monoterpenoid indole alkaloid biosynthesis. In: Romeo JT, Saunders JA, Matthews BF (eds) Recent advances in phytochemistry: regulation of phytochemicals by molecular techniques, Vol 35. Elsevier Science, Oxford, pp 275–295
Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 5:733–739
Holmberg CI, Tran SE, Eriksson JE, Sistonen L (2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem Sci 27:619–627
Irmler S, Schröder G, St-Pierre B, Crouch NP, Hotze M, Schmidt J, Strack D, Matern U, Schröder J (2000) Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J 24:797–804
Jensen AB, Raventos D, Mundy J (2002) Fusion genetic analysis of jasmonate-signalling mutants in Arabidopsis. Plant J 29:595–606
Luijendijk TJC, van der Meijden E, Verpoorte R (1996) Involvement of strictosidine as a defensive chemical in Catharanthus roseus. J Chem Ecol 22:1355–1366
Lopes Cardoso MI, Meijer AH, Rueb S, Queiroz Machado J, Memelink J, Hoge JHC (1997) A promoter region that controls basal and elicitor-inducible expression levels of the NADPH: cytochrome P450 reductase gene (Cpr) from Catharanthus roseus binds nuclear factor GT-1. Mol Gen Genet 256:674–681
Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950
McKnight TD, Roessner CA, Devagupta R, Scott AI, Nessler CL (1990) Nucleotide sequence of a cDNA encoding the vacuolar protein strictosidine synthase from Catharanthus roseus. Nucleic Acids Res 18:4939
Meijer AH, Verpoorte R, Hoge JHC (1993a) Regulation of enzymes and genes involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J Plant Res 3:145–164
Meijer AH, Lopes Cardoso MI, Voskuilen JT, de Waal A, Verpoorte R, Hoge JHC (1993b) Isolation and characterization of a cDNA clone from Catharanthus roseus encoding NADPH: cytochrome P-450 reductase, an enzyme essential for reactions catalysed by cytochrome P-450 mono-oxygenases in plants. Plant J 4:47–60
Memelink J, Verpoorte R, Kijne JW (2001a) ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci 6:212–219
Memelink J, Kijne JW, van der Heijden R, Verpoorte R (2001b) Genetic modification of plant secondary metabolite pathways using transcriptional regulators. In: Scheper T (ed) Advances in biochemical engineering/biotechnology, Vol 72. Springer-Verlag, Berlin Heidelberg, pp 103–125
Menke FLH, Parchmann S, Mueller MJ, Kijne JW, Memelink J (1999a) Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in Catharanthus roseus. Plant Physiol 119:1289–1296
Menke FLH, Champion A, Kijne JW, Memelink J (1999b) A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor- inducible AP2-domain transcription factor, ORCA2. EMBO J 18:4455–4463
Moreno PRH, van der Heijden R, Verpoorte R (1995) Cell and tissue cultures of Catharanthus roseus: a literature survey II. Updating from 1988–1993. Plant Cell Tiss Org Cult 42:1–25
Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13:1959–1968
Ouwerkerk PBF, Hallard D, Verpoorte R, Memelink J (1999a) Identification of UV-B light-responsive regions in the promoter of the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol 41:491–503
Ouwerkerk PBF, Trimborn TO, Hilliou F, Memelink J (1999b) Nuclear factors GT-1 and 3AF1 interact with multiple sequences within the promoter of the Tdc gene from Madagascar periwinkle: GT-1 is involved in UV light-induced expression. Mol Gen Genet 261:610–622
Ouwerkerk PBF, Memelink J (1999a) A G-box element from the Catharanthus roseus strictosidine synthase (Str) gene promoter confers seed-specific expression in transgenic tobacco plants. Mol Gen Genet 261:635–643
Ouwerkerk PBF, Memelink J (1999b) Elicitor-responsive promoter regions in the tryptophan decarboxylase gene from Catharanthus roseus. Plant Mol Biol 39:129–136
Pasquali G, Goddijn OJM, de Waal A, Verpoorte R, Schilperoort RA, Hoge JHC, Memelink J (1992) Coordinated regulation of two indole alkaloid biosynthetic genes from Catharanthus roseus by auxin and elicitors. Plant Mol Biol 18:1121–1132
Pasquali G, Erven ASW, Ouwerkerk PBF, Menke FLH, Memelink J (1999) The promoter of the strictosidine synthase gene from periwinkle confers elicitor-inducible expression in transgenic tobacco and binds nuclear factors GT-1 and GBF. Plant Mol Biol 39:1299–1310
Pauw B, Hilliou FAO, Sandonis Martin V, Chatel G, de Wolf CJF, Champion A, Pré M, van Duijn B, Kijne JW, van der Fits L, Memelink J (2004) Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J Biol Chem 279:52940–52948
Pauw B, Memelink J (2005) Jasmonate-responsive gene expression. J Plant Growth Regul 23:200–210
Pré M, Sibéril Y, Memelink J, Champion A, Doireau P, Gantet P (2000) Isolation by the yeast one-hybrid system of cDNAs encoding transcription factors that bind to the G-box element of the strictosidine synthase gene promoter from Catharanthus roseus. Int J Bio-chrom 5:229–244
Rijhwani SK, Shanks JV (1998) Effect of elicitor dosage and exposure time on biosynthesis of indole alkaloids by Catharanthus roseus hairy root cultures. Biotechnol. Prog 14:442–449
Roewer IA, Cloutier C, Nessler CL, De Luca V (1992) Transient induction of tryptophan decarboxylase (TDC) and strictosidine synthase (SS) genes in cell suspension cultures of Catharanthus roseus. Plant Cell Rep 11:86–89
Rojo E, Titarenko E, Leon J, Berger S, Vancanneyt G, Sanchez-Serrano JJ (1998) Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J 13:153–165
Schröder G, Unterbusch E, Kaltenbach M, Schmidt J, Strack D, De Luca V, Schröder J (1999) Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett 458:97–102
Sibéril Y, Benhamron S, Memelink J, Giglioli-Guivarc’h N, Thiersault M, Boisson B, Doireau P, Gantet P (2001) Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol Biol 45:477–488
Sottomayor M, Ros Barcelo A (2003) Peroxidase from Catharanthus roseus (L.) G. Don and the biosynthesis of α-3′,4′-anhydrovinblastine: a specific role for a multifunctional enzyme. Protoplasma 222:97–105
Sottomayor M, Lopes Cardoso I, Pereira LG, Ros Barceló A (2004) Peroxidase and the biosynthesis of terpenoid indole alkaloids in the medicinal plant Catharanthus roseus (L.) G. Don Phytochem Rev 3:159–171
St-Pierre B, Laflamme P, Alarco AM, De Luca V (1998) The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J 14:703–713
van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289:295–297
van der Fits L, Memelink J (2001) The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J 25:43–53
van der Fits L, Zhang H, Menke FLH, Deneka M, Memelink J (2000) A Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a jasmonate-independent signal transduction pathway. Plant Mol Biol 44:675–685
van der Fits L, Hilliou F, Memelink J (2001) T-DNA activation tagging as a tool to isolate regulators of a metabolic pathway from a genetically non-tractable plant species. Transgenic Res 10:513–521
Vazquez-Flota F, De Carolis E, Alarco AM, De Luca V (1997) Molecular cloning and characterization of desacetoxyvindoline-4-hydroxylase, a 2-oxoglutarate dependent-dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L.) G. Don Plant Mol Biol 34:935–948
Vazquez-Flota FA, De Luca V (1998) Jasmonate modulates development- and light-regulated alkaloid biosynthesis in Catharanthus roseus. Phytochemistry 49:395–402
Vazquez-Flota F, Carrillo-Pech M, Minero-Garcia Y, De Lourdes Miranda-Ham M (2004) Alkaloid metabolism in wounded Catharanthus roseus seedlings. Plant Physiol Biochem 42:623–628
Vom Endt D (2004) Regulation of jasmonate-responsive ORCA transcription factors in Catharanthus roseus. PhD Thesis, Leiden University, The Netherlands
Vom Endt D, Kijne JW, Memelink J (2002) Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry 61:107–114
Acknowledgements
The collaboration between the authors was financially supported by a European Van Gogh 2000–2001 exchange grant (ref 00106NH).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Memelink, J., Gantet, P. Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus . Phytochem Rev 6, 353–362 (2007). https://doi.org/10.1007/s11101-006-9051-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11101-006-9051-z