Abstract
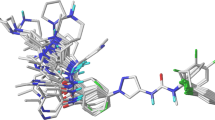
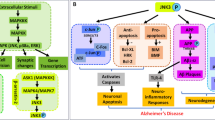
Within the mitogen-activated protein kinase (MAPK) 1 family, the two kinases of c-Jun N-terminal kinase 3 (JNK3) and p38α MAPK have emerged in the last decades as particularly attractive therapeutic targets due to their implication in several neurodegenerative pathologic conditions. In this study, the structure and activity relationship of 60 dual JNK3/p38α MAPK inhibitors was explored; three-dimensional quantitative structure-activity relationship (3D-QSAR), including comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis fields (CoMSIA), was performed. From the data we got, the 3D-QSAR model (CoMFAJNK3 with q2 is 0.642, r2 is 0.958; CoMSIAJNK3 with q2 is 0.660, r2 is 0.963; CoMFAp38α with q2 is 0.605, r2 is 0.980; CoMSIAp38α with q2 is 0.608, r2 is 0.970) had good predictability. Molecular docking further revealed the binding mode of inhibitors to JNK3/p38α MAPK. The results of 3D-QSAR, molecular docking, and molecular dynamics simulation also provided guidance for the discovery of new dual inhibitors of JNK3 and p38α MAPK. Finally, 10 novel compounds with good potential activity and ADME/T profile were designed. Molecular dynamics simulation results validated that Met149/Lys 93/Gln 155 (JNK3) and Met109/Lys53 (p38α) located in the active site play a key role for novel dual inhibitors.








Similar content being viewed by others
References
Fricker M, LoGrasso P, Ellis S, Wilkie N, Hunt P, Pollack SJ (2005) Substituting c-Jun N-terminal kinase-3 (JNK3) ATP-binding site amino acid residues with their p38 counterparts affects binding of JNK- and p38-selective inhibitors. Arch Biochem Biophys 438:195–205. https://doi.org/10.1016/j.abb.2005.04.013
Resnick L, Fennell M (2004) Targeting JNK3 for the treatment of neurodegenerative disorders. Drug Discov Today 9:932–939. https://doi.org/10.1016/S1359-6446(04)03251-9
Messoussi A, Feneyrolles C, Bros A, Deroide A, Daydé-Cazals B, Chevé G (2014) Recent progress in the design, study, and development of c-Jun N-terminal kinase inhibitors as anticancer agents. Chem Biol 21:1433-1443. https://doi.org/10.1016/j.chembiol.2014.09.007
Mielke K, Herdegen T (2000) JNK and p38 stresskinases-degenerative effectors of signal-transduction-cascades in the nervous system. Prog Neurobiol 61:45–60. https://doi.org/10.1016/S0301-0082(99)00042-8
Ansideri F, Macedo JT, Eitel M, El-Gokha A, Zinad DS, Scarpellini C, et al (2018) Structural optimization of a pyridinylimidazole scaffold: shifting the selectivity from p38α mitogen-activated protein kinase to c-Jun N-terminal kinase 3. ACS Omega 3:7809-7831. https://doi.org/10.1021/acsomega.8b00668
Koch P, Jahns H, Schattel V, Goettert M, Laufer S (2010) Pyridinylquinoxalines and pyridinylpyridopyrazines as lead compounds for novel p38α mitogen-activated protein kinase inhibitors. J Med Chem 53:1128–1137. https://doi.org/10.1021/jm901392x
Muth F, El-Gokha A, Ansideri F, Eitel M, Döring E, Sievers-Engler A et al (2017) Tri- and tetrasubstituted pyridinylimidazoles as covalent inhibitors of c-Jun N-terminal kinase 3. J Med Chem 60:594–607. https://doi.org/10.1021/acs.jmedchem.6b01180
Muth F, Günther M, Bauer SM, Döring E, Fischer S, Maier J et al (2015) Tetra-substituted pyridinylimidazoles as dual inhibitors of p38α mitogen-activated protein kinase and c-Jun N-terminal kinase 3 for potential treatment of neurodegenerative diseases. J Med Chem 58:443–456. https://doi.org/10.1021/jm501557a
Ansideri F, Lange A, El-Gokha A, Boeckler FM, Koch P (2016) Fluorescence polarization-based assays for detecting compounds binding to inactive c-Jun N-terminal kinase 3 and p38α mitogen-activated protein kinase. Anal Biochem 503:28–40. https://doi.org/10.1016/j.ab.2016.02.018
Fu L, Chen Y, C-m X, Wu T, H-m G, Lin Z-h et al (2020) 3D-QSAR, HQSAR, molecular docking, and new compound design study of 1,3,6-trisubstituted 1,4-diazepan-7-ones as human KLK7 inhibitors. Med Chem Res 29:1012–1029. https://doi.org/10.1007/s00044-020-02542-3
Clark M, Cramer RD, Jones DM, Patterson DE, Simeroth PE (1990) Comparative molecular field analysis (CoMFA). 2. Toward its use with 3D-structural databases. Tetrahedron Comput Methodol 3:47–59. https://doi.org/10.1016/0898-5529(90)90120-W
Klebe G, Abraham UJJoC-AMD (1999) Comparative molecular similarity index analysis (CoMSIA) to study hydrogen-bonding properties and to score combinatorial libraries. J Comput Aided Mol Des 13:1–10. https://doi.org/10.1023/a:1008047919606
Bush B, Nachbar RJJCAMD (1993) Sample-distance partial least squares: PLS optimized for many variables, with application to CoMFA. J Comput Aided Mol Des 7:587–619. https://doi.org/10.1007/bf00124364
Wendt B, Cramer RJJCAMD (2014) Challenging the gold standard for 3D-QSAR: template CoMFA versus X-ray alignment. J Comput Aided Mol Des 28:803–824. https://doi.org/10.1007/s10822-014-9761-z
Golbraikh A, Tropsha A (2002a) Beware of q2! J Mol Graph Model 20:269–276. https://doi.org/10.1016/s1093-3263(01)00123-1
Golbraikh A, Tropsha AJJCAMD (2002b) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput Aided Mol Des 16:357–369. https://doi.org/10.1023/a:1021372108686
Mitra I, Roy PP, Kar S, Ojha PK, Roy KJJC (2010) On further application of r2m as a metric for validation of QSAR models. J Chemom 24:22–33. https://doi.org/10.1002/cem.1268
Pratim Roy P, Paul S, Mitra I, Roy KJM (2009) On two novel parameters for validation of predictive QSAR models. Molecules. 14:1660–1701. https://doi.org/10.3390/molecules14051660
Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J Chem Theory Comput 8:1542–1555. https://doi.org/10.1021/ct200909j
Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput 9:3878–3888. https://doi.org/10.1021/ct400314y
Sprenger KG, Jaeger VW, Pfaendtner J (2015) The General AMBER Force Field (GAFF) Can accurately predict thermodynamic and transport properties of many ionic liquids. J Phys Chem B 119:5882–5895. https://doi.org/10.1021/acs.jpcb.5b00689
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO et al (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct Funct Bioinformatics 78:1950–1958. https://doi.org/10.1002/prot.22711
Sun H, Duan L, Chen F, Liu H, Wang Z, Pan P et al (2018) Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys Chem Chem Phys 20:14450–14460. https://doi.org/10.1039/c7cp07623a
Huang K, Luo S, Cong Y, Zhong S, Zhang JZH, Duan L (2020) An accurate free energy estimator: based on MM/PBSA combined with interaction entropy for protein–ligand binding affinity. Nanoscale. 12:10737–10750. https://doi.org/10.1039/c9nr10638c
Ertl P, Schuffenhauer A (2009) Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J Cheminformatics 1:8. https://doi.org/10.1186/1758-2946-1-8
Abdizadeh T, Ghodsi R, Hadizadeh F (2017) 3D-QSAR (CoMFA, CoMSIA) and molecular docking studies on histone deacetylase 1 selective inhibitors. Recent Pat Anti-Cancer Drug Discov 12:365–383. https://doi.org/10.2174/1574892812666170508125927
Astolfi A, Kudolo M, Brea J, Manni G, Manfroni G, Palazzotti D et al (2019) Discovery of potent p38α MAPK inhibitors through a funnel like workflow combining in silico screening and in vitro validation. Eur J Med Chem 182:111624. https://doi.org/10.1016/j.ejmech.2019.111624
Laufer SA, Hauser DRJ, Domeyer DM, Kinkel K, Liedtke AJ (2008) Design, synthesis, and biological evaluation of novel tri- and tetrasubstituted imidazoles as highly potent and specific ATP-mimetic inhibitors of p38 MAP kinase: focus on optimized interactions with the enzyme’s surface-exposed front region. J Med Chem 51:4122–4149. https://doi.org/10.1021/jm701529q
Funding
The authors are grateful to the Natural Science Foundation of China (81171508), the Key Project of Chongqing Natural Science Foundation (cstc2015jcyjBX0080), and the Scientific Research Startup Fund of Chongqing University of Technology (2017ZD42).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 2356 kb)
Rights and permissions
About this article
Cite this article
Fu, L., Chen, Y., Guo, Hm. et al. A selectivity study of polysubstituted pyridinylimidazoles as dual inhibitors of JNK3 and p38α MAPK based on 3D-QSAR, molecular docking, and molecular dynamics simulation. Struct Chem 32, 819–834 (2021). https://doi.org/10.1007/s11224-020-01668-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01668-9