Abstract
X-ray analysis of N-(4-fluorophenyl)-1,5-dimethyl-1H-imidazole-4-carboxamide 3-oxide reveals the temperature-dependent polymorphism associated with the crystallographic symmetry conversion. The observed crystal structure transformation corresponds to a symmetry reduction from I41 /a (I) to P43 (II) space groups. The phase transition mainly concerns the subtle but clearly noticeable reorganization of molecules in the crystal space, with the structure of individual molecules left almost unchanged. The Hirshfeld surface analysis shows that various intermolecular contacts play an important role in the crystal packing, revealing graphically the differences in spatial arrangements of the molecules in both polymorphs. The N-oxide oxygen atom acts as a formally negatively charged hydrogen bonding acceptor in intramolecular hydrogen bond of N–H…O− type. The combined crystallographic and theoretical DFT methods demonstrate that the observed intramolecular N-oxide N–H…O hydrogen bond should be classified as a very strong charge-assisted and closed-shell non-covalent interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Research on new imidazole and benzimidazole N-oxides and their applications in biology has focused particular interest in recent years [1]. Some of them exhibit biological activity, e.g., as insecticides, plant growth regulators, antiinflammatory, and antiprotozoal agents [1–3]. More importantly, imidazole N-oxides are highly attractive intermediates for the preparation of diverse polyfunctionalized imidazole-based compounds of biological significance. For example, a series of protein kinase inhibitors [4] was synthesized by the so-called “sulfur transfer reaction” [5], whereas direct palladium-catalyzed arylation protocol opened up direct access to potent Tie2 tyrosine kinase inhibitor [6]. Special attention was paid to the synthesis of imidazole N-oxides with new substitution patterns, particularly the analogs lacking a substituent at carbon C(2) atom, and their transformations into more complex derivatives [5, 7–9]. A large number of key 2-unsubstituted N-oxides exhibit limited stability either under high temperature, UV irradiation or in the presence of acylating agents, and can undergo isomerization to the corresponding imidazol-2-ones. Therefore, we turned our attention to derivatives bearing hydrogen donor groups at the vicinal C(4) position, namely amide [10, 11] and hydrazide [12, 13] moieties. The presence of such groups and stable N-oxide function offers an opportunity for their application in more complex structures including biologically active compounds and enables carrying out the reactions under harsh conditions. Some reports on imidazole derivatives containing amide or hydrazide groups as potential pharmaceuticals were also published recently [14].
In this paper, we report the results of X-ray crystal structure determination of N-(4-fluorophenyl)-1,5-dimethyl-1H-imidazole-4-carboxamide 3-oxide polymorphs (Scheme 1). The title compound shows the temperature-dependent polymorphism resulting from a rearrangement of the molecules in the crystal due to temperature variations. A comparative characterization of both polymorphs was performed, and details concerning structural differences between the polymorphic forms were discussed in the context of intermolecular interactions present in the crystal state. In addition, the result of temperature-dependent X-ray measurement was presented in order to highlight the details of the observed phase transition. Intermolecular non-covalent interactions were studied by means of Hirshfeld surface approach.
Methods
Synthesis and crystallization
The title compound was prepared according to the known protocol [15–17] by cyclocondensation of a respective N-aryl α-hydroxyimino-β-oxobutyramide and 1,3,5-trimethylhexahydro-1,3,5-triazine in aqueous ethanol. The synthesis was performed starting with commercially available ethyl acetoacetate, paraformaldehyde, methylamine (40 % aqueous solution), and 4-fluoroaniline. All other reagents and solvents were purchased and used as received without further purification. The crystals suitable for X-ray analysis were obtained by slow evaporation of the solvent from an ethanol solution.
X-ray structure determination
X-ray diffraction measurements were made on a four-circle Oxford Diffraction Xcalibur diffractometer equipped with a two-dimensional area CCD detector with the graphite monochromatized MoKα radiation and a low-temperature device Cryostream cooler Oxford Cryosystem. Integration of the intensities and correction for Lorentz and polarization effects were performed using the CrysAlis RED software [18].
The crystal structures were solved by direct methods using the SHELXS program [19]. The appropriate choice of a space group was based on the following tests: analysis of Wilson plot and the distributions of normalized structure factors; checking Laue symmetry; and the analysis of systematic absences (details are given in the supplement). The following space groups were proposed as the most adequate candidate space groups: the centrosymmetric I41/a tetragonal space group for room-temperature data (293 K), and the non-centrosymmetric P43 one for the low-temperature data (150 K). With this choice of space groups, the use of direct methods provided proper tentative crystal structure models (positions of all the non-hydrogen atoms were found in both cases). As both polymorphic structures are very similar to each other, some problems could appear concerning the correctness of space group assignment and the obtained crystal structure models. The key problem here is to explain if the observed differences are not due to erroneous data reduction, inappropriate space group choice, or other errors. To eliminate such a situation, we made some attempts at finding crystal structure models in several space groups (for details see supporting information file associated with this paper). However, apart from the above-mentioned space groups, other space groups occurred to be interchangeably inadequate. Detailed comparison of polymorphic structures is given in section “Temperature-dependent study of polymorphism.”
The crystal structures were then refined by a full-matrix least-squares method on F 2 using the SHELXL-97 program [20] implemented in WinGX [21] suite of programs. The positions of NH hydrogen atoms were found on Fourier difference map and refined. Hydrogen atoms of the aromatic rings and methyl groups were introduced in the calculated positions with idealized geometry. They were constrained using a rigid body model with isotropic displacement parameters equal to 1.2 or 1.5 of the equivalent displacement parameters of the parent atoms. For the appropriate modeling of methyl groups, disorder in (I) two groups of hydrogen atoms were introduced and constrained in staggered geometry (positions rotated relative each other by 60° and occupation factors were fixed at 0.5 for each atom). Details of both refinements are presented in Table 1.
A summary of relevant crystallographic data is given in Table 1. The molecular geometry was calculated by PARST [22] and Platon [23]. Selected bond distances and angles are summarized in Table 2. Atomic coordinates, displacement parameters, and structure factors are deposited with Cambridge Crystallographic Data Centre CCDC.Footnote 1
Theoretical computations
Theoretical quantum chemical calculations were performed with the Gaussian 09 sets of codes [24]. The molecular geometry taken from the X-ray studies was fully optimized. For this purpose, DFT-B3LYP functional was used in conjunction with 6-311++G(d,p) basis set. Further, QTAIM calculations were done with the use of AIMAll program [25].
Hirshfeld surface analysis
The Hirshfeld molecular surfaces and the associated fingerprint plots were generated using CrystalExplorer 3.0 [26, 27] on the basis of X-ray results. The bonds of hydrogen atoms were normalized to standard neutron values (C–H = 1.083 Å, O–H = 0.983 Å, N–H = 1.009 Å) [28]. For comparison of intermolecular interactions in the crystal structures, the Hirshfeld surfaces were mapped with normalized contact distances (d norm). The D norm parameter is based on d i (the distance from the surface to the nearest atom in the molecule itself), d e (the distance from the surface to the nearest atom in another molecule), and van der Waals radii of the corresponding atoms (r vdWi and r vdWe ). It is given by the Eq. (1).
All the presented graphical plots use the same red–white–blue color scheme, where the red color highlights the shortest intermolecular atomic contacts (negative d norm values), white is used for contacts around the van der Waals separation, and blue corresponds to longer ones (positive d norm values). The Hirshfeld surface fingerprint plots were generated using d i and d e as a pair of coordinates at intervals of 0.01 Å. A color gradient in the plots ranging from blue to red represents the proportional contribution of contact pairs in the global surface.
Results and discussion
Temperature-dependent study of polymorphism
On the basis of NMR spectroscopic data [11], it was expected that there exists a relatively strong intramolecular hydrogen bond between N-oxide and N–H amide groups. Our preliminary room-temperature X-ray results unambiguously confirmed this suggestion. The molecular structure of the title compound is presented in Fig. 1. Using the same crystal, we additionally collected a new diffraction data set at low temperature (150 K) in order to investigate the behavior of proton within the intramolecular N–H…O hydrogen bridge. Surprisingly, the comparison of X-ray structure determination indicated polymorphic transformation upon the change of temperature.
Molecular drawings of the title compound determined by X-ray analysis: a room-temperature structure (I); b low-temperature structure (II) with a representative molecule IIA. Atomic displacement ellipsoids are drawn at the 50 % probability level. Intramolecular N–H…O hydrogen bonds are presented as dashed lines (Color figure online)
The room-temperature polymorph (I) crystallizes in the centrosymmetric I41/a tetragonal space group with one molecule in a general position. However, this model of the crystal structure proved to be completely inadequate for refinements against low-temperature diffraction data. For an appropriate crystal structure determination, we had to establish a new structural model. The crystal structure of polymorph (II) was solved by direct methods and refined successfully in the non-centrosymmetric P43 space group with four independent molecules in the asymmetric unit (IIA, IIB, IIC, and IID). There were only small differences in geometrical parameters when these four molecules were compared (Table 2).
The title polymorphic structures can be distinguished one from another when their crystal packing was analyzed in detail. At both temperatures, the crystal structures are composed of two kinds of molecular layers alternating along the crystallographic c direction (Fig. 2). In each layer, the molecules are roughly parallel to each other due to stacking interactions. The presence of these layers results from the crystallographic four-fold screw axes and from the particular position of molecules with respect to those symmetry elements. The differences between polymorphs can be revealed when comparing how the molecules in a layer are arranged in relation to the crystallographic directions. In (II), the molecules in layers are parallel to each other but evidently twisted with respect to the crystallographic c direction as opposed to (I). The dihedral angle between the main molecular plane (calculated through positions of all non-hydrogen atoms) and (001) crystallographic plane is equal to 89.6(2)° in (I) and varies from 76.6(2)° to 78.9(2)° in (II). The observed differences in a mutual molecular arrangement in each separate layer are presented in Fig. 3.
Crystal packing revealing the layered nature of the polymorphic structures: room-temperature structure in I41/a space group (I) (a); low-temperature structure in P43 space group (II) (b). View perpendicular to a direction. Direction of a selected four-fold screw axis is presented with blue color in both unit cells (Color figure online)
Moreover, while the choice of a space group for (I) raises no doubts, some controversies may arise in the case of (II) because of four symmetrically independent molecules in the asymmetric unit. Figure 4 shows that the temperature transformation is certainly connected with the change of a space group because of evident loss of (½, ½, and ½) translation characteristic of the I unit cell. Hence, according to the International Tables for Crystallography [29] in the same crystal system in a primitive unit cell, the only possible choice of a standard setting space group is P43.
Interestingly, taking the four crystallographic independent molecules of (II) into account, it was observed that their conformations are similar in pairs IIA/IID and IIB/IIC and different between these pairs. There are pseudo-inversion centers between the molecular couples, which may be compared with crystallographic inversion centers in (I). Also, a pseudo glide plane can be observed which is perpendicular to the c direction originating from the crystallographic symmetry glide plane a z in the polymorph (I).
Summing up, upon cooling from 293 to 150 K, the crystal underwent spontaneous phase transition assisted by subtle structural effects of molecular rearrangement leading to the lowering of the crystallographic symmetry. It seems that the new low-temperature molecular rearrangement is mainly associated with the reduction of volumes occupied by the molecules in the structure of polymorph (II). However, the structural transformation is revealed primarily by an anomalous reduction of c-axis length in comparison with the other two axes. The relative shortenings of lattice parameters upon the change of temperature can be described by Δd/d = [(d 293K − d 150K)/d 293K (d corresponds to a lattice parameter under consideration). Hence, Δd/d is equal to 0.018(1) for the c lattice parameter, and it is much smaller 0.003(1) for the a lattice parameter. While comparing both polymorphs, there are also different V/Z proportions which change from 303.6(3)Å3 for (I) to 296.7(2)Å3 for (II).
In order to analyze the effects associated with the phase transition, a systematic study of the dependence of lattice parameters on temperature was performed. The X-ray diffraction experiment, based on typical measurements for the unit cell determination, was carried out at temperatures ranging from 300 to 145 K with a 5 K step change. In the temperature range of 240–160 K, a problem appeared with indexing reflections in the tetragonal system and only the monoclinic unit cell was found (Table 3). Nevertheless, X-ray measurements for crystal structure determination at 230 K afforded successful crystal structure solution only in the triclinic P − 1 space group (V/Z = 300.6 Å3). The results of structure determination based on these data are of rather poor quality indicating dynamic behavior of the molecules upon crystal cooling. In turn, X-ray measurements below 160 K led to the tetragonal unit cell of polymorph (II). It was then postulated that the intermediate triclinic form represents a phase of transition state between two tetragonal polymorphic structures.
In the temperature range of 240–160 K, the molecules exhibit large reorientation motions in the solid state resulting in significant distortions from mean atomic positions. As explained by Ulrich [30], the molecular displacements may start in one or several cells at the same time but then they are followed by other cells. Hence, these processes, although they take place in the solid state, resemble nucleation and growth of a new crystal phase. In the described case, the molecular reorganization starts upon decreasing the temperature to about 240 K and is completed below 160 K.
It may thus be concluded that by means of temperature-dependent X-ray diffraction, we observed that a single crystal, the object of our studies, underwent an order–disorder–order phase transition from one tetragonal form (I) to the other (II) by intermediate triclinic structure. This transformation results from a dynamic molecular rearrangement in the crystal structure leading to two distinct molecular patterns. Interestingly, upon warming of the crystal to room temperature again structure (I) is recovered. This regularity in the changes of the crystal structure allowed us to classify the observed phase transition as a reversible non-reconstructive one, assisted by lowering crystal point symmetry [31]. Similar crystal structure transformations are more common for inorganic compounds [32]. It is also known that among phase transitions induced by the change of temperature, the highest temperature modification as a rule has higher symmetry [30].
It is worth pointing out that, as it will also be demonstrated, the phase transition results mostly from the changes in crystal packing. In turn, the structural differences observed for individual molecules are meaningless in practice, which is described in detail in section “Molecular structure”.
Molecular structure
The molecular structures (I) and (II) are very similar. Small discrepancies in the molecular conformations are best described by the torsion angles including the atoms O1, N1, C5, C6, N7, and C8. However, the corresponding angle values, as listed in Table 2, do not differ by more than 5°. In all the examined cases, the values of C5–C6–N7–C8 torsion angles clearly confirmed the existence of molecular trans conformations in central amide linkage. Interestingly, the room-temperature studies revealed a disorder of hydrogen atoms of both methyl groups in an imidazole ring. In contrast to that, no such disorder was observed for the low-temperature results; hence, this phenomenon may also be related to the polymorphism.
The N-(4-fluorophenyl)-1,5-dimethyl-1H-imidazole-4-carboxamide 3-oxide molecule can be divided into three individual chemical fragments: a nitrone-like N-oxidoimidazole part, a fluorine substituted phenyl ring, and an amide moiety linking both aromatic systems. The dihedral angles between phenyl and imidazole range from 10.5(1)° to 11.7(1)° in both polymorphs. In turn, the dihedral angles of the imidazole rings and the amide planes are much smaller (between 3.3(1)° and 5.5(1)°).
In order to describe the overall molecular shape, the least-squares mean planes through positions of all non-hydrogen atoms were calculated for each of the molecules. In all the cases, the molecules could be best described as almost planar with the maximum atomic deviation from the main molecular plane of about 0.2 Å.
The arrangement of single-double bonds in the molecule and almost coplanar positions of the atoms forming the main molecular skeleton both make possible π-communication between the aromatic rings, and therefore affect the lengths of some covalent bonds. A particularly strong interaction was observed for internal C–N and C–C bonds of the imidazole rings. For example, the lengths of N1–C2 and C4–C5 bonds varied from 1.303(3)Å to 1.354(3)Å and 1.347(3)Å to 1.397(3)Å, respectively, in the analyzed molecules. In turn, C4–C5 bond lengths ranged from 1.464(3)Å to 1.487(3)Å. Similarly, N1–O1 N-oxide bond differentiation was also observed.
The observed small differences in geometrical parameters between (I) and (II) (Table 3) can be related to different refining strategies—refinement in different space groups with Z’ = 1 (I) or Z’ = 4 (II) results in a different data/parameter ratio. In general, the bond lengths observed for (I) better resemble literature values [28].
Intramolecular hydrogen bond
In both polymorphs, molecular trans conformations are stabilized by intramolecular N7–H7…O1 hydrogen bonds (Fig. 1). These interactions, closing six-membered rings, can be formally classified as charge-assisted ones [33, 34]. The original reason of the temperature X-ray studies (293 and 150 K) was related to the comparative determination of atom positions within N7–H7…O1 hydrogen bridges. The relatively long N–H bond and a large value of N–H…O bond angle in (I) allow one to classify this intramolecular hydrogen bond as a strong one. Moreover, the short N…O distance suggests that it could also be considered as a low-barrier hydrogen bond [35, 36] similar to intramolecular hydrogen bonds observed for benzopyrane derivatives [37–39]. To elucidate this observation, low-temperature X-ray measurements were undertaken. However, hydrogen bonding geometry obtained from those studies, did not show any significant differences in comparison with room-temperature results, except for a little higher N–H…O angle values (Table 4 and Figure S1 in the Supplementary Material). Therefore, for a more detailed analysis of intramolecular hydrogen bonds, we used the methods of quantum chemistry.
In order to estimate the approximate interaction energy in the investigated intramolecular N–H…O bridge, we used the topological electron density approach. According to this method, the hydrogen bonding energy can be calculated by equation Eint = −1/2VH-BCP in which VH-BCP is the electron potential energy density measured in the hydrogen bond critical bond [40]. In view of the fact that the hydrogen atoms positions derived from X-ray measurements were uncertain, the geometry of the molecule taken from the crystal structure was fully optimized at the B3LYP/6-311++G** level, and then the QTAIM analysis was performed [41]. Selected geometrical and QTAIM parameters obtained via theoretical calculations are collected in Table 5.
The obtained interaction energy is equal to 12.1 kcal/mol. Therefore, the QTAIM analysis confirmed the predictions made on the basis of the X-ray structural properties that the intramolecular N7–H7…O1 contact can be classified as a strong hydrogen bond, especially when compared with the binding energy of hydrogen bonds in water dimer, which is about 4–5 kcal/mol [42]. As already mentioned, the investigated hydrogen bond can be classified as a charge-assisted one and it has been demonstrated recently that for this type of interactions the energy obtained from QTAIM analysis may be underestimated due to the fact that electrostatic interactions are not reflected directly by the charge distribution [43, 44]. Thus, it is possible that the N7–H7…O1 contact is in fact much stronger than it could result from a direct relation between VH-BCP and Eint. Nevertheless, the positive value of the Laplacian of electron density calculated in H…O BCP suggests that the closed-shell character of the N7–H7…O1 interaction is maintained, even if this interaction is relatively stronger than a typical hydrogen bond of moderate strength. The structural manifestation of the relatively high-hydrogen bonding interaction energy is also evident as the N7–H7 bond elongation, simultaneous N7…O1 distance shortening and an almost linear arrangement of N7, H7, and O1 atoms are obtained from the X-ray studies of both polymorphs.
Intermolecular interactions
Despite the observed temperature-dependent change of the crystallographic space group, the low-temperature structure (II) in general resembles the room-temperature one (I). In both cases, there are two kinds of molecular layers alternating along crystallographic c direction, stabilized by stacking interactions. These π…π interactions affect the arrangement of adjacent imidazole and phenyl rings. The distances between imidazole rings of the neighboring molecules, changing from 3.32(1)Å to 3.47(1)Å, are associated with centroids offset of about 1.3Å. In turn, the distances between phenyl rings range from 3.36(1)Å to 3.70(1)Å with larger offsets of about 2.7Å.
The molecular layers are connected to each other by a network of intermolecular C–H…O hydrogen bonds. The N-oxide O1 atom as an acceptor takes part in two kinds of hydrogen bonding: N7–H7…O1 intramolecular bonding and C2–H2…O1 intermolecular one. As a result of the latter interaction (linking neighboring imidazole rings), a molecular tetramer around inversion 4-fold axis is formed (Fig. 5). According to the graph-set notation [45], it can be designated by the R 44 (16) first-level motif. In turn, the C13-H13…O6 interaction between phenyl rings and carbonyl groups, orders molecules into C(4) chains extending along the c axis. Small discrepancies between (I) and (II) in the corresponding hydrogen bonding geometric parameters are presented in Table 5.
Hirshfeld surface analysis
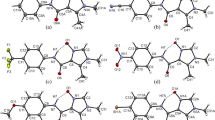
The observed polymorphism turned our attention to the behavior of particular atoms as the centers of intermolecular interactions. For a detailed analysis of the interactions scheme, we calculated molecular Hirshfeld surfaces (Fig. 6). The Hirshfeld surface of (I) is based on all observed orientations of disordered methyl groups including their partial occupancies. Obviously, the surfaces obtained for the investigated molecules resemble each other in shape, volume, and area (Table 6). It seems that the differences in the volumes are mainly caused by the change of temperature.
Mapping d norm on Hirshfeld surfaces not only allows one to decode dominant intermolecular interactions in the crystal structures but also presents subtle differences in the scheme of intermolecular contacts. On the surfaces calculated for each separate molecule, there are seen regions of intermolecular C–H…O hydrogen bonds as the most intensive large red circles near the oxygen and C–H hydrogen atoms. There are also many short intermolecular contacts involving fluorine and hydrogen atoms. The corresponding red circles occur in the vicinity of fluorine atoms.
The right side of the Fig. 6 presents Hirshfeld surface fingerprint plots. The dominant interactions between atoms of neighboring molecules are shown as the bright areas colored with light blue and green. A very useful thing is that the fingerprint plots can be decomposed to highlight the particular atom pairs in close intermolecular contacts. Decomposition of individual types of intermolecular contacts, based on Hirshfeld surface fingerprint plots, indicates the areas of H…F contacts. As the positions of hydrogen atoms were constrained, detailed discussion on these intermolecular contacts is not included. On the other hand, a high proportional contribution of H…F contacts on all the surfaces should be noticed here. In the fingerprint plots, dominant intermolecular interactions (bright colored areas) are attributed mainly to the C–H…O hydrogen bonds. These interactions are represented by typical long- sharp spikes of minimum d e values below 1.2 Å. Interestingly, in the case of (I), there is also a central spike (about 1.2 Å) resulting from short H…H contacts.
The percentage of various intermolecular contacts in the Hirshfeld surfaces is presented in Table 7. The relatively high proportion of C/N/O…C/N/O contacts (around 10 %) is attributed to π…π stacking interactions. The contributions of different intermolecular contacts can be treated as indicators of discrepancies between the investigated molecules as interacting neighbors in the crystal state. Hence, for (II) there is a growing contribution of intermolecular contacts associated with stacking interactions.
The above results of Hirshfeld surface analysis clearly show the changes in crystal packing, in particular when comparing fingerprint plots estimated for individual molecules present in both polymorphs.
Conclusions
The results of X-ray structure determination revealed molecular rearrangement in the title crystal upon change of conditions. The observed phenomenon is defined as a reversible order–disorder–order phase conversion associated with the lowering of crystallographic symmetry.
In both polymorphs, the observed molecular trans conformations are stabilized by intramolecular N–H…O hydrogen bonds. These interactions were analyzed with the help of QTAIM approach and turned out to be very strong charge-assisted hydrogen bonds.
The use of the Hirshfeld surface analysis provided an interesting insight into the crystal structure as it is seen by the interacting neighboring molecules. Our studies showed that, besides the C–H…O intermolecular hydrogen bonds and π…π stacking interactions there is also a significant contribution of H…F intermolecular contacts in each of the investigated structures. The comparison of fingerprint plots estimated for individual molecules gives graphical evidence of the difference in crystal packing found for both polymorphic phases.
Notes
The supplementary crystallographic data for this paper (932320, 932321 and 943140) can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html or from the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax: +44-1223-336033.
References
Begtrup M (2012) In: Diazole, Triazole, and Tetrazole N-Oxides. Adv Heterocycl Chem 106:37; and the references cited therein
Cerecetto H, González M (2002) Current Topic Med Chem 2:1187
Aguirre G, Boiani M, Cerecetto H, Gerpe A, González M, Sainz YF, Denicola A, de Ocáriz CO, Nogal JJ, Montero D, Escario JA (2004) Arch Pharm Pharm Med Chem 337:259
Dimova D, Iyer P, Vogt M, Totzke F, Kubbutat MHG, Schächtele C, Laufer S, Bajorath J (2012) J Med Chem 55:11067
Campeau LC, Stuart DR, Leclerc JP, Bertrand-Laperle M, Villemure E, Sun HY, Lasserre S, Guimond N, Lecavallier M, Fagnou K (2009) J Am Chem Soc 131:3291
Mlostoń G, Gendek T, Heimgartner H (1998) Helv Chim Acta 81:1585
Mlostoń G, Jasiński M, Linden A, Heimgartner H (2006) Helv Chim Acta 89:1304
Mlostoń G, Romański J, Jasiński M, Heimgartner H (2009) Tetrahedron: Asymmetry 20:1073
Mlostoń G, Jasiński M, Heimgartner H, Eur J Organ Chem (2011) 2542
Jasiński M, Mlostoń G, Linden A, Heimgartner H (2008) Helv Chim Acta 91:1916
Mlostoń G, Jasiński M (2010) Collect Czech Chem Commun 75:871
Mlostoń G, Pieczonka AM, Kowalczyk E, Linden A, Heimgartner H (2011) Helv Chim Acta 94:1764
Pieczonka AM, Mlostoń G, Heimgartner H (2012) Helv Chim Acta 95:404
Lange JHM, van Stuivenberg HH, Coolen HKAC, Adolfs TJP, McCreary AC, Keizer HG, Wals HC, Veerman W, Borst AJM, de Looff W, Verveer PC, Kruse CG (2005) J Med Chem 48:1823
Hadizadeh F, Hosseinzadeh H, Sadat Motamed-Shariaty V, Seifi M, Kazemi S (2008) Iran J Pharm Res 7:29
Pellei M, Gandin V, Marinelli M, Marzano C, Yousufuddin M, Dias HVR, Santini C (2012) Inorg Chem 51:9873
Chae E, Shin YJ, Ryu EJ, Ji MK, Cho NR, Lee KH, Jeong HJ, Kim SJ, Choi Y, Oh KS, Park CE, Yoon YS (2013) Bioorg Med Chem Lett 23:2134
Oxford Diffraction (2008) CrysAlis CCD and CrysAlis RED. Versions 1.171.32.29 Oxford Diffraction Ltd, Abingdon, England
Sheldrick GM (1986) SHELXS86 Program for Crystal Structure Solution. University of Göttingen, Germany
Sheldrick GM (2008) Acta Cryst A64:112
Farrugia LJ (1999) J Appl Cryst 32:837
Nardelli M (1995) J Appl Cryst 28:659
Spek AL (2009) Acta Cryst D65:148
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE et al (2009) Gaussian 09 (Revision A.02), Gaussian, Inc., Wallingford, CT
Keith TA, AIMAll Version 09.11.29, 2009 (aim.tkgristmill.com)
Spackman MA, McKinnon JJ, Jayatilaka D (2008) CrystEngComm 10:377
Spackman MA, Jayatilaka D (2009) CrystEngComm 11:19
Allen FH, Kennard O, Watson DG, Brammer L, Orpen AG, Taylor R (1987) J Chem Soc Perkin Trans 2:S1
International Tables for Crystallography (2006) Part A, Chapter 3.1. Kluwer, Dordrecht, the Netherlands
Ulrich M (2008) Symmetry Relations between Crystal Structures. Summer School on Mathmatical and Thoeretical Crystallography, Gargano, Italy
Tolédano JC, Janovec V, Kopský V, Scott JF, Boček P (2006) International Tables for Crystallography, Part D, Chapter 3.1. Kluwer, Dordrecht, the Netherlands
Bujak M, Angel RJ (2005) J Solid State Chem 178:2237
Gilli P, Gilli G (2000) J Mol Struct 552:1
Gilli P, Bertolasi V, Pretto L, Gilli G (2006) J Mol Struct 790:40
Frey PA, Whitt SA, Tobin JB (1994) Science 264:1927
Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM (1997) J Am Chem Soc 119:1081
Rybarczyk A, Olszak TA, Małecka M, Nawrot-Modranka J (1999) Acta Crystallogr C55:1313
Rybarczyk-Pirek AJ, Małecka M, Grabowski SJ, Nawrot-Modranka J (2002) Acta Crystallogr C58:o405
Rybarczyk-Pirek AJ, Dubis AT, Grabowski AJ, Nawrot-Modranka J (2006) Chem Phys 320:247
Espinosa E, Molins E, Lecomte C (1998) Chem Phys Lett 285:170
Bader RFW (1990) Atoms in Molecules: A Quantum Theory. University Press, New York, Oxford
Jabłoński M, Palusiak M (2012) J Phys Chem A 116:2322
Bankiewicz B, Palusiak M (2011) Comput Theor Chem 966:113
Bankiewicz B, Matczak P, Palusiak M (2012) J Phys Chem A 116:452
Etter MC, MacDonald JC, Bernstein J (1990) Acta Crystallogr Sect B 46:256
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Rybarczyk-Pirek, A.J., Łukomska, M., Ejsmont, K. et al. Temperature-dependent polymorphism of N-(4-fluorophenyl)-1,5-dimethyl-1H-imidazole-4-carboxamide 3-oxide: experimental and theoretical studies on intermolecular interactions in the crystal state. Struct Chem 25, 979–989 (2014). https://doi.org/10.1007/s11224-014-0404-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-014-0404-8