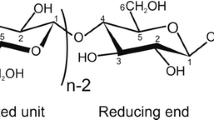
Abstract
The result of DFT geometry optimization of 68 unique α-maltose conformers at the B3LYP/6-311++G** level of theory is described. Particular attention is paid to the hydroxyl group rotational positions and their influence on the glycosidic bond dihedral angles. The orientation of lone pair electrons across the bridging hydrogen bonds are implicated in directing the glycosidic dihedral angles with for example, conformers gg-gg and gt-gt, having different minimum energy conformations for the clockwise (c) and the reverse clockwise (r) forms. Conformers tg-gg, gg-tg, tg-tg, gt-gg, and gg-gt were studied, to understand the intermediate glycosidic bond conformations. The conformation, tg-gg-c, was found to be the lowest energy structure. When the hydroxyl groups on each glucose residue were made to point in opposite directions, i.e., c/r and r/c, the optimized structures were found to have high relative energies. Several optimized ‘kink’ structures were found around (\(\phi_{\rm H}\), ψH) ∼(−40°, −40°), the lowest relative energy conformation being ∼3 kcal/mol. “Kink” conformations are observed in crystalline CA-10 and CA-14mers. Band-flip conformations, also observed in X-ray structures of CA-26 fragments, were studied with the lowest energy α-maltose conformations ∼4.0 kcal/mol above the global energy minimum. Several trends in geometry resulting from hydroxyl rotamer directions are described.










Similar content being viewed by others
References
Momany FA, Willett JLJ (2000) Comput Chem 21:1204
Strati GL et al (2002) Carbohydr Res 337:1833
Strati GL et al (2002) Carbohydr Res 337:1851
Bosma WB et al (2006) J Mol Struct:THEOCHEM 776:1
Bosma WB et al (2006) J Mol Struct:THEOCHEM 776:13
Appell M et al (2004) Carbohydr Res 339:537
Momany FA et al (2006) Carbohydr Res 341:525
Appell M et al (2005) Carbohydr Res 340:459
Schnupf U et al (2007) Carbohydr Res 342:196
Momany FA et al (2005) Carbohydr Res 340:1638
Momany FA et al (2004) Carbohydr Res 339:553
Umemura M et al (2005) J Mol Struct: THEOCHEM 730:1
Lins RD, Hunenberger PH (2005) J Comput Chem 26:1400
Kony D et al (2004) J Chem Phys B 108:5815
Stortz CA, Cerezo AS (2003) Carbohydr Res 338:95
Cheetham NWH et al (2003) Carbohydr Res 338:955
Kuttel M et al (2002) J Comput Chem 23:1236
Mendonca SJ et al (2002) Phys Chem A 106:4115
Engelsen SB et al (2001) Biophys Chem 93:103
French AD et al (2001) J Comput Chem 22:65
Momany FA, Willett JL (2000) Carbohydr Res 326:194
French AD et al (2000) J Mol Graph Model 18:95
French AD et al (2000) J Mol Struct 556:303
Spieser SAH et al (1999) Carbohydr Res 322:264
Simmerling C et al (1998) J Amer Chem Soc 120:5771
Damm W et al (1997) J Comput Chem 18:1955
Sakurai M et al (1997) Bull Chem Soc Jpn 70:847
Senderowitz H et al (1997) J Org Chem 62:1427
Ott K-H, Meyer BJ (1996) Carbohydr Res 381:11; (1996) J Comput Chem 17:1068
Reiling S et al (1996) J Comput Chem 17:450
Woods RJ et al (1995) J Phys Chem 99:3832
Schmidt RK et al (1995) J Phys Chem 99:11339
Fringant C et al (1995) Carbohydr Res 278:27
Kouwijzer MLCE, Grootenhuis PDJ (1995) J Phys Chem 99:13426
Glennon TM et al (1994) J Comput Chem 15:1019
Brady JW, Schmidt RK (1993) J Phys Chem 97:958
French AD, Dowd MK (1993) J Mol Struct: THEOCHEM 286:183
Dowd MK et al (1992) Carbohydr Res 230:223
French AD, Brady JW (1990) In: French AD, Brady JW (eds) Computer modeling of carbohydrate molecules, ACS Symposium Series 430. American Chemical Society, Washington, DC, p 1
Rasmussen K, Frabricius J (1990) In: French AD, Brady JW (eds) Computer modeling of carbohydrate molecules, ACS Symposium Series 430. American Chemical Society, Washington, DC, p 177
French AD (1989) Carbohydr Res 188:206
Tran V et al (1989) Biopolymers 28:679
Ha SN et al (1988) Carbohydr Res 180:207
Ha SN et al (1988) Biopolymers 27:1927
Koehler JEH et al (1987) Eur Biophys J 15:197
Shaskov AS et al (1986) Carbohydr Res 147:175
Tvaroska I, Perez S (1986) Carbohydr Res 149:389
Lipkind GM et al (1984) Carbohydr Res 133:1
Melberg S, Rasmussen K (1979) Carbohydr Res 69:27; (1980) Carbohydr Res 78:215
Sugiyama H et al (2000) Carbohydr Res 325:177
Shimada J et al (2000) J Phys Chem B 104:2136
Pangborn W et al (1985) Int J Biol Macromol 7:363
Taga T et al (1993) Carbohydr Res 240:39
Taga T et al (1994) Carbohydr Res 251:203
Jeffrey GA, Huang D (1991) Carbohydr Res 222:47
Quigley GJ et al (1970) J Am Chem Soc 92:5834
Gres SME, Jeffrey GA (1977) Acta Crystallogr B33:2490
Takusagawa F, Jacobson RA (1978) Acta Crystallogr B34:213
Chu SSC, Jeffrey GA (1967) Acta Crystallogr 23:1038
Tanaka I et al (1976) Acta Crystallogr B32:155
Imberty A, Perez S (1988) Carbohydr Res 181:41
Brisse F et al (1982) J Am Chem Soc 104:7470
Saenger W et al (1998) Chem Rev 98:1787
Nimz O et al. (2001) Carbohydr Res 336:141
Stevens ES, Sathyanarayana BK (1989) J Amer Chem Soc 111:4149
Stevens ES (1992) Biopolymers 32:1571
Insight/Discover, Molecular Simulations Inc, 9685 Scranton Road, San Diego, CA 92121-3752
Becke AD (1993) J Chem Phys 98:5648
For the performance of B3LYP with a split valence basis set, see (a) Novoa JJ, Sosa CJ (1995) Phys Chem 99:15837; Sirois S et al (1997) J Chem Phys 107:6770; Paizs B, Suhai S (1998) J Comput Chem 19:575; Hagemeister FC et al (1998) J Phys Chem A 102:82
PQS Ab Initio Program Package, Parallel Quantum Solutions, 2013 Green Acres, Suite E, Fayetteville, AR 72703, USA
HyperChem 7.5, Hypercube, Inc, 115 NW 4th Street, Gainsville, FL 32601 USA
Newton MD, Jeffrey GA, Takagi S (1979) J Amer Chem Soc 101:1997
Suzuki T et al (2006) J Phys Chem B 110:2405
Tvaroska I et al (2002) Carbohydr Res 337:353
Dyekjaer JD, Rassmussen K (2003) Mini Rev Med Chem 3:713
Author information
Authors and Affiliations
Corresponding author
Additional information
Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Momany, F.A., Schnupf, U., Willett, J.L. et al. DFT study of α-maltose: influence of hydroxyl orientations on the glycosidic bond. Struct Chem 18, 611–632 (2007). https://doi.org/10.1007/s11224-007-9191-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-007-9191-9